
ChatCell: Facilitating Single-Cell Analysis with Natural Language
💻Demo • 🤗 Dataset • 🏖️ Overview • 🧬 Single-cell Analysis Tasks • 🛠️ Quickstart • 📝 Cite
📌 Table of Contents
🛠️ Quickstart
from transformers import AutoTokenizer, AutoModelForSeq2SeqLM
tokenizer = AutoTokenizer.from_pretrained("zjunlp/chatcell-large")
model = AutoModelForSeq2SeqLM.from_pretrained("zjunlp/chatcell-large")
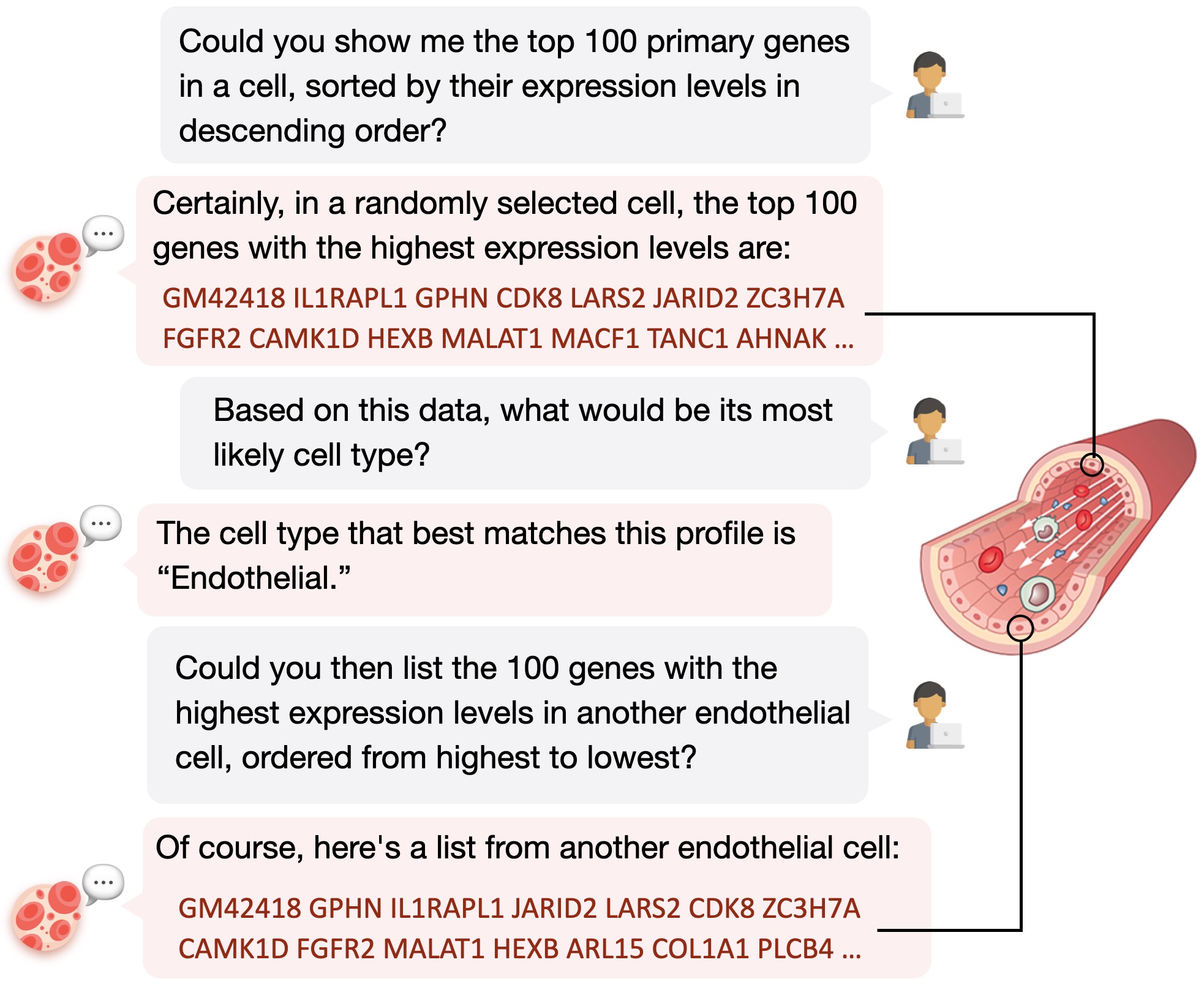
input_text="Detail the 100 starting genes for a Mix, ranked by expression level: "
# Encode the input text and generate a response with specified generation parameters
input_ids = tokenizer(input_text,return_tensors="pt").input_ids
output_ids = model.generate(input_ids, max_length=512, num_return_sequences=1, no_repeat_ngram_size=2, top_k=50, top_p=0.95, do_sample=True)
# Decode and print the generated output text
output_text = tokenizer.decode(output_ids[0],skip_special_tokens=True)
print(output_text)
🧬 Single-cell Analysis Tasks
ChatCell can handle the following single-cell tasks:
- Random Cell Sentence Generation. Random cell sentence generation challenges the model to create cell sentences devoid of predefined biological conditions or constraints. This task aims to evaluate the model's ability to generate valid and contextually appropriate cell sentences, potentially simulating natural variations in cellular behavior.

- Pseudo-cell Generation. Pseudo-cell generation focuses on generating gene sequences tailored to specific cell type labels. This task is vital for unraveling gene expression and regulation across different cell types, offering insights for medical research and disease studies, particularly in the context of diseased cell types.

- Cell Type Annotation. For cell type annotation, the model is tasked with precisely classifying cells into their respective types based on gene expression patterns encapsulated in cell sentences. This task is fundamental for understanding cellular functions and interactions within tissues and organs, playing a crucial role in developmental biology and regenerative medicine.

- Drug Sensitivity Prediction. The drug sensitivity prediction task aims to predict the response of different cells to various drugs. It is pivotal in designing effective, personalized treatment plans and contributes significantly to drug development, especially in optimizing drug efficacy and safety.

📝 ✨ Acknowledgements
Special thanks to the authors of Cell2Sentence: Teaching Large Language Models the Language of Biology and Representing cells as sentences enables natural-language processing for single-cell transcriptomics for their inspiring work.
📝 Cite
@article{fang2024chatcell,
title={ChatCell: Facilitating Single-Cell Analysis with Natural Language},
author={Fang, Yin and Liu, Kangwei and Zhang, Ningyu and Deng, Xinle and Yang, Penghui and Chen, Zhuo and Tang, Xiangru and Gerstein, Mark and Fan, Xiaohui and Chen, Huajun},
year={2024},
}
- Downloads last month
- 18
This model does not have enough activity to be deployed to Inference API (serverless) yet. Increase its social
visibility and check back later, or deploy to Inference Endpoints (dedicated)
instead.