|
--- |
|
license: apache-2.0 |
|
datasets: |
|
- nferruz/UR50_2021_04 |
|
tags: |
|
- chemistry |
|
- biology |
|
--- |
|
|
|
|
|
### Model Description |
|
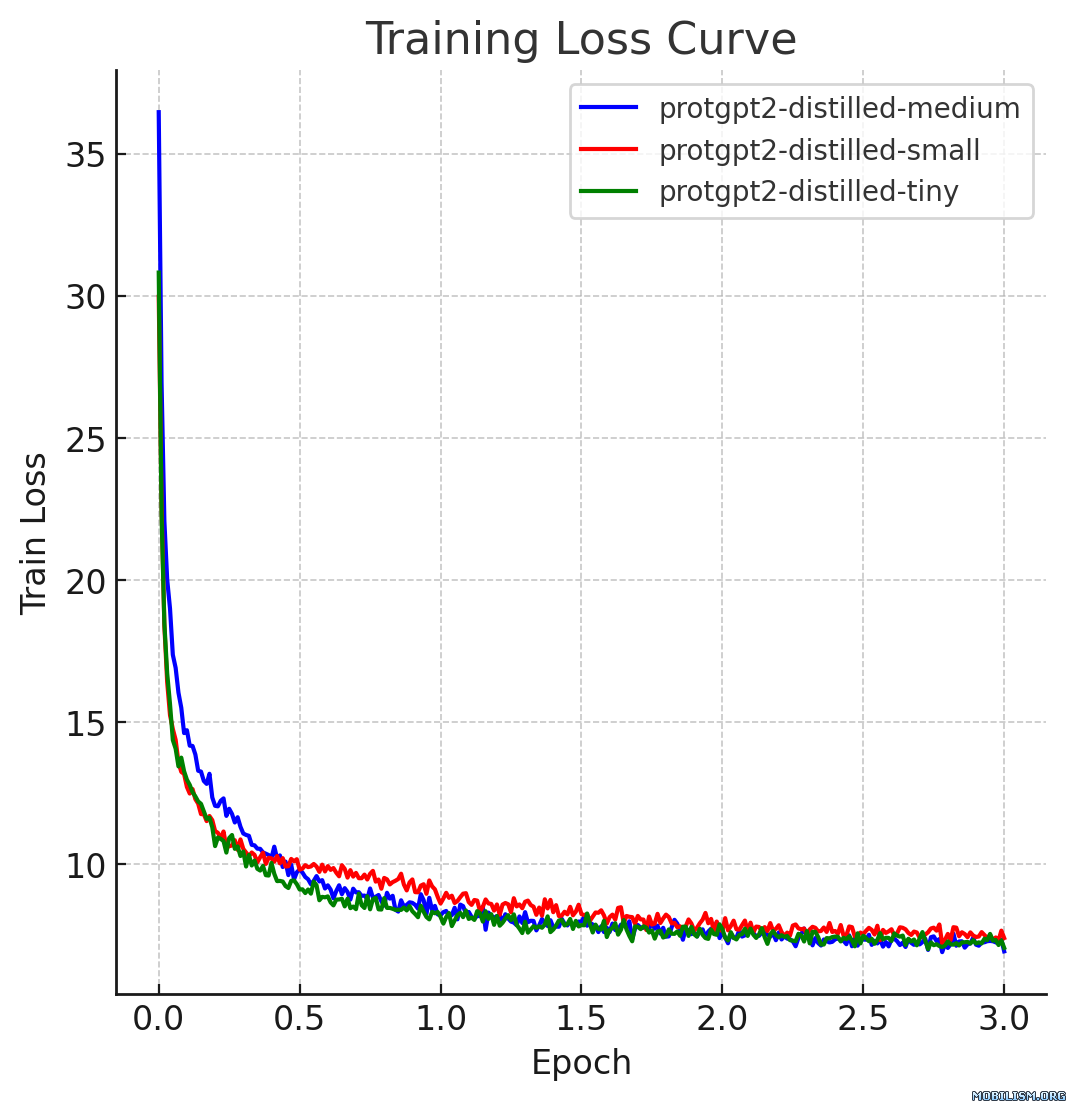
This model card describes the distilled version of [ProtGPT2](https://huggingface.co./nferruz/ProtGPT2), referred to as `protgpt2-distilled-tiny`. The distillation process for this model follows the methodology of knowledge distillation from a larger teacher model to a smaller, more efficient student model. The process combines both "Soft Loss" (Knowledge Distillation Loss) and "Hard Loss" (Cross-Entropy Loss) to ensure the student model not only generalizes like its teacher but also retains practical prediction capabilities. |
|
|
|
### Technical Details |
|
**Distillation Parameters:** |
|
- **Temperature (T):** 10 |
|
- **Alpha (α):** 0.1 |
|
- **Model Architecture:** |
|
- **Number of Layers:** 4 |
|
- **Number of Attention Heads:** 4 |
|
- **Embedding Size:** 512 |
|
|
|
**Dataset Used:** |
|
- The model was distilled using a subset of the evaluation dataset provided by [nferruz/UR50_2021_04](https://huggingface.co./datasets/nferruz/UR50_2021_04). |
|
|
|
<strong>Loss Formulation:</strong> |
|
<ul> |
|
<li><strong>Soft Loss:</strong> <span>ℒ<sub>soft</sub> = KL(softmax(s/T), softmax(t/T))</span>, where <em>s</em> are the logits from the student model, <em>t</em> are the logits from the teacher model, and <em>T</em> is the temperature used to soften the probabilities.</li> |
|
<li><strong>Hard Loss:</strong> <span>ℒ<sub>hard</sub> = -∑<sub>i</sub> y<sub>i</sub> log(softmax(s<sub>i</sub>))</span>, where <em>y<sub>i</sub></em> represents the true labels, and <em>s<sub>i</sub></em> are the logits from the student model corresponding to each label.</li> |
|
<li><strong>Combined Loss:</strong> <span>ℒ = α ℒ<sub>hard</sub> + (1 - α) ℒ<sub>soft</sub></span>, where <em>α</em> (alpha) is the weight factor that balances the hard loss and soft loss.</li> |
|
</ul> |
|
<p><strong>Note:</strong> KL represents the Kullback-Leibler divergence, a measure used to quantify how one probability distribution diverges from a second, expected probability distribution.</p> |
|
|
|
|
|
|
|
### Performance |
|
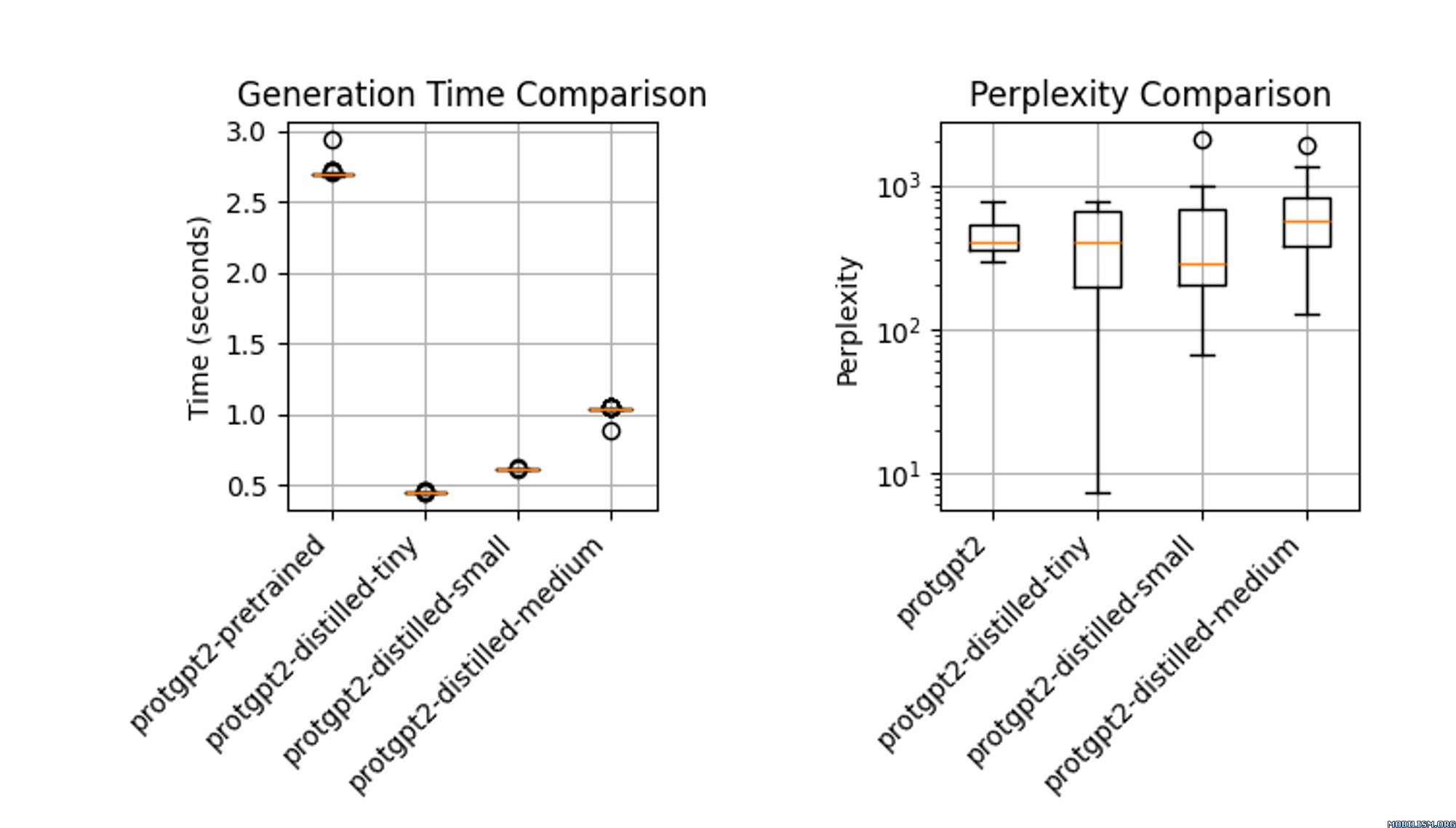
The distilled model, `protgpt2-distilled-tiny`, demonstrates a substantial increase in inference speed—up to 6 times faster than the pretrained version. This assessment is based on evaluations using \(n=100\) tests, showing that while the speed is significantly enhanced, the model still maintains perplexities comparable to the original. |
|
|
|
 |
|
|
|
|
|
 |
|
|
|
### Usage |
|
|
|
``` |
|
from transformers import GPT2Tokenizer, GPT2LMHeadModel, TextGenerationPipeline |
|
import re |
|
|
|
# Load the model and tokenizer |
|
model_name = "littleworth/protgpt2-distilled-tiny" |
|
tokenizer = GPT2Tokenizer.from_pretrained(model_name) |
|
model = GPT2LMHeadModel.from_pretrained(model_name) |
|
|
|
# Initialize the pipeline |
|
text_generator = TextGenerationPipeline( |
|
model=model, tokenizer=tokenizer, device=0 |
|
) # specify device if needed |
|
|
|
# Generate sequences |
|
generated_sequences = text_generator( |
|
"<|endoftext|>", |
|
max_length=100, |
|
do_sample=True, |
|
top_k=950, |
|
repetition_penalty=1.2, |
|
num_return_sequences=10, |
|
pad_token_id=tokenizer.eos_token_id, # Set pad_token_id to eos_token_id |
|
eos_token_id=0, |
|
truncation=True, |
|
) |
|
|
|
def clean_sequence(text): |
|
# Remove the "<|endoftext|>" token |
|
text = text.replace("<|endoftext|>", "") |
|
|
|
# Remove newline characters and non-alphabetical characters |
|
text = "".join(char for char in text if char.isalpha()) |
|
|
|
return text |
|
|
|
# Print the generated sequences |
|
for i, seq in enumerate(generated_sequences): |
|
cleaned_text = clean_sequence(seq["generated_text"]) |
|
print(f">Seq_{i}") |
|
print(cleaned_text) |
|
``` |
|
|
|
### Use Cases |
|
1. **High-Throughput Screening in Drug Discovery:** The distilled ProtGPT2 facilitates rapid mutation screening in drug discovery by predicting protein variant stability efficiently. Its reduced size allows for swift fine-tuning on new datasets, enhancing the pace of target identification. |
|
2. **Portable Diagnostics in Healthcare:** Suitable for handheld devices, this model enables real-time protein analysis in remote clinical settings, providing immediate diagnostic results. |
|
3. **Interactive Learning Tools in Academia:** Integrated into educational software, the distilled model helps biology students simulate and understand protein dynamics without advanced computational resources. |
|
|
|
### References |
|
- Hinton, G., Vinyals, O., & Dean, J. (2015). Distilling the Knowledge in a Neural Network. arXiv:1503.02531. |
|
- Original ProtGPT2 Paper: [Link to paper](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9329459/) |