MoLFormer-XL-both-10%
MoLFormer is a class of models pretrained on SMILES string representations of up to 1.1B molecules from ZINC and PubChem. This repository is for the model pretrained on 10% of both datasets.
It was introduced in the paper Large-Scale Chemical Language Representations Capture Molecular Structure and Properties by Ross et al. and first released in this repository.
Model Details
Model Description
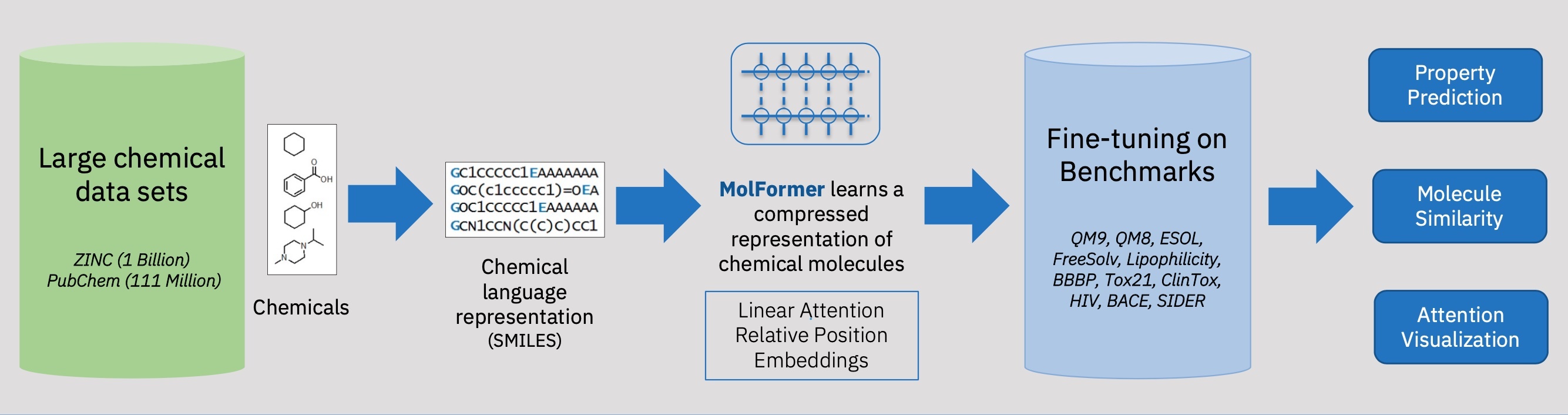
MoLFormer is a large-scale chemical language model designed with the intention of learning a model trained on small molecules which are represented as SMILES strings. MoLFormer leverges masked language modeling and employs a linear attention Transformer combined with rotary embeddings.
An overview of the MoLFormer pipeline is seen in the image above. One can see that the transformer-based neural network model is trained on a large collection of chemical molecules represented by SMILES sequences from two public chemical datasets PubChem and ZINC in a self-supervised fashion. The MoLFormer architecture was designed with an efficient linear attention mechanism and relative positional embeddings with the goal of learning a meaningful and compressed representation of chemical molecules. After training the MoLFormer foundation model was then adopted to different downstream molecular property prediction tasks via fine-tuning on task-specific data. To further test the representative power of MoLFormer, the MoLFormer encodings were used to recover molecular similarity, and analysis on the correspondence between the interatomic spatial distance and attention value for a given molecule was performed.
Intended use and limitations
You can use the model for masked language modeling, but it is mainly intended to be used as a feature extractor or to be fine-tuned for a prediction task. The "frozen" model embeddings may be used for similarity measurements, visualization, or training predictor models. The model may also be fine-tuned for sequence classification tasks (e.g., solubility, toxicity, etc.).
This model is not intended for molecule generation. It is also not tested for molecules larger than ~200 atoms (i.e., macromolecules). Furthermore, using invalid or noncanonical SMILES may result in worse performance.
Example code
Use the code below to get started with the model.
import torch
from transformers import AutoModel, AutoTokenizer
model = AutoModel.from_pretrained("ibm/MoLFormer-XL-both-10pct", deterministic_eval=True, trust_remote_code=True)
tokenizer = AutoTokenizer.from_pretrained("ibm/MoLFormer-XL-both-10pct", trust_remote_code=True)
smiles = ["Cn1c(=O)c2c(ncn2C)n(C)c1=O", "CC(=O)Oc1ccccc1C(=O)O"]
inputs = tokenizer(smiles, padding=True, return_tensors="pt")
with torch.no_grad():
outputs = model(**inputs)
outputs.pooler_output
Training Details
Data
We trained MoLFormer-XL on a combination of molecules from the ZINC15 and PubChem datasets. This repository contains the version trained on 10% ZINC + 10% PubChem.
Molecules were canonicalized with RDKit prior to training and isomeric information was removed. Also, molecules longer than 202 tokens were dropped.
Hardware
- 16 x NVIDIA V100 GPUs
Evaluation
We evaluated MoLFormer by fine-tuning on 11 benchmark tasks from MoleculeNet. The tables below show the performance of different MoLFormer variants:
| BBBP | HIV | BACE | SIDER | ClinTox | Tox21 | |
|---|---|---|---|---|---|---|
| 10% ZINC + 10% PubChem | 91.5 | 81.3 | 86.6 | 68.9 | 94.6 | 84.5 |
| 10% ZINC + 100% PubChem | 92.2 | 79.2 | 86.3 | 69.0 | 94.7 | 84.5 |
| 100% ZINC | 89.9 | 78.4 | 87.7 | 66.8 | 82.2 | 83.2 |
| MoLFormer-Base | 90.9 | 77,7 | 82.8 | 64.8 | 61.3 | 43.1 |
| MoLFormer-XL | 93.7 | 82.2 | 88.2 | 69.0 | 94.8 | 84.7 |
| QM9 | QM8 | ESOL | FreeSolv | Lipophilicity | |
|---|---|---|---|---|---|
| 10% ZINC + 10% PubChem | 1.7754 | 0.0108 | 0.3295 | 0.2221 | 0.5472 |
| 10% ZINC + 100% PubChem | 1.9093 | 0.0102 | 0.2775 | 0.2050 | 0.5331 |
| 100% ZINC | 1.9403 | 0.0124 | 0.3023 | 0.2981 | 0.5440 |
| MoLFormer-Base | 2.2500 | 0.0111 | 0.2798 | 0.2596 | 0.6492 |
| MoLFormer-XL | 1.5984 | 0.0102 | 0.2787 | 0.2308 | 0.5298 |
We report AUROC for all classification tasks, average MAE for QM9/8, and RMSE for the remaining regression tasks.
Citation
@article{10.1038/s42256-022-00580-7,
year = {2022},
title = {{Large-scale chemical language representations capture molecular structure and properties}},
author = {Ross, Jerret and Belgodere, Brian and Chenthamarakshan, Vijil and Padhi, Inkit and Mroueh, Youssef and Das, Payel},
journal = {Nature Machine Intelligence},
doi = {10.1038/s42256-022-00580-7},
pages = {1256--1264},
number = {12},
volume = {4}
}
@misc{https://doi.org/10.48550/arxiv.2106.09553,
doi = {10.48550/ARXIV.2106.09553},
url = {https://arxiv.org/abs/2106.09553},
author = {Ross, Jerret and Belgodere, Brian and Chenthamarakshan, Vijil and Padhi, Inkit and Mroueh, Youssef and Das, Payel},
keywords = {Machine Learning (cs.LG), Computation and Language (cs.CL), Biomolecules (q-bio.BM), FOS: Computer and information sciences, FOS: Computer and information sciences, FOS: Biological sciences, FOS: Biological sciences},
title = {Large-Scale Chemical Language Representations Capture Molecular Structure and Properties},
publisher = {arXiv},
year = {2021},
copyright = {arXiv.org perpetual, non-exclusive license}
}
- Downloads last month
- 228,558