MSAGPT
MSAGPT📖 Paper: MSAGPT: Neural Prompting Protein Structure Prediction via MSA Generative Pre-Training MSAGPT is a powerful protein language model (PLM). MSAGPT has 3 billion parameters with three versions of the model, MSAGPT, MSAGPT-Sft, and MSAGPT-Dpo, supporting zero-shot and few-shot MSA generation. MSAGPT achieves state-of-the-art structural prediction performance on natural MSA-scarce scenarios. |
Overall Framework

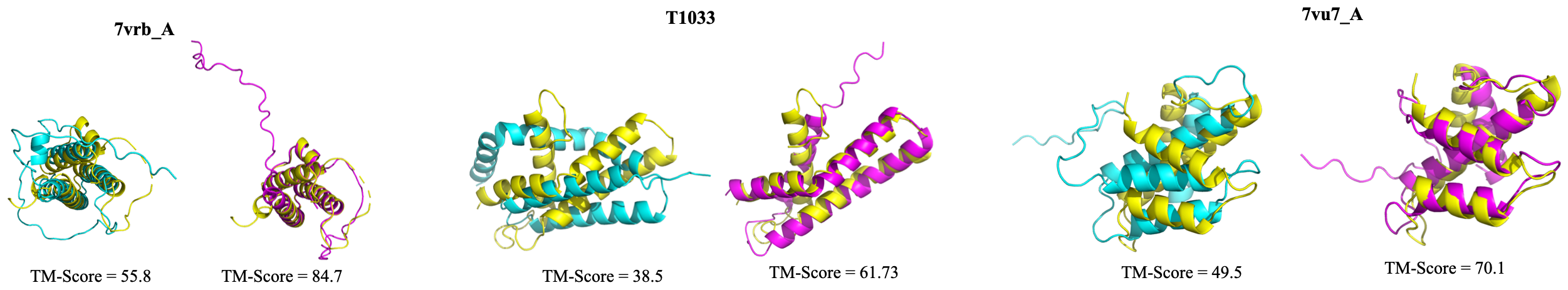
Visualized Cases
Visualization of improved structure prediction compared with nature MSA. Yellow: Ground truth; Purple: Predictions based on MSA generated by MSAGPT; Cyan: Predictions from MSA generated by natural MSA.
Get Started:
Option 1:Deploy MSAGPT by yourself
We support GUI for model inference.
First, we need to install the dependencies.
# CUDA >= 11.8
pip install -r requirements.txt
Model List
You can choose to manually download the necessary weights. Then UNZIP it and put it into the checkpoints folder.
| Model | Type | Seq Length | Download |
|---|---|---|---|
| MSAGPT | Base | 16K | 🤗 Huggingface 🔨 SwissArmyTransformer |
| MSAGPT-SFT | Sft | 16K | 🤗 Huggingface 🔨 SwissArmyTransformer |
| MSAGPT-DPO | Rlhf | 16K | 🤗 Huggingface 🔨 SwissArmyTransformer |
Situation 1.1 CLI (SAT version)
Run CLI demo via:
# Online Chat
bash scripts/cli_sat.sh --from_pretrained ./checkpoints/MSAGPT-DPO --input-source chat --stream_chat --max-gen-length 1024
The program will automatically interact in the command line. You can generate replies entering the protein sequence you need to generate virtual MSAs (or add a few MSAs as a prompt, connected by "<M>"), for example: "PEGKQGDPGIPGEPGPPGPPGPQGARGPPG<M>VTVEFVNSCLIGDMGVDGPPGQQGQPGPPG", where "PEGKQGDPGIPGEPGPPGPPGPQGARGPPG" is the main sequence, and "VTVEFVNSCLIGDMGVDGPPGQQGQPGPPG" are MSA prompts, and pressing enter. Enter stop to stop the program. The chat CLI looks like:

You can also enable the offline generation by set the --input-source <your input file> and --output-path <your output path>. We set an input file example: msa_input.
# Offline Generation
bash scripts/cli_sat.sh --from_pretrained ./checkpoints/MSAGPT-DPO --input-source <your input file> --output-path <your output path> --max-gen-length 1024
Situation 1.2 CLI (Huggingface version)
(TODO)
Situation 1.3 Web Demo
(TODO)
Option 2:Finetuning MSAGPT
(TODO)
Hardware requirement
Model Inference: For BF16: 1 * A100(80G)
Finetuning:
For BF16: 4 * A100(80G) [Recommend].
License
The code in this repository is open source under the Apache-2.0 license.
If you find our work helpful, please consider citing the our paper
@article{chen2024msagpt,
title={MSAGPT: Neural Prompting Protein Structure Prediction via MSA Generative Pre-Training},
author={Chen, Bo and Bei, Zhilei and Cheng, Xingyi and Li, Pan and Tang, Jie and Song, Le},
journal={arXiv preprint arXiv:2406.05347},
year={2024}
}