library_name: tf-keras
license: mit
pipeline_tag: image-classification
tags:
- cancer detection
- histopathological image classification
- breast cancer diagnose
- breast cancer detection
- umair akram
Breast Cancer Detection using CNNs in TensorFlow
In this project, a Convolutional Neural Network (CNN) is employed for the purpose of detecting Breast Cancer. The algorithm takes patches of Histopathological Images of patients' breast tissues and utilizes CNNs to ascertain whether the breast tissues within the image patch contain Invasive Ductal Carcinoma (IDC) or not. By analyzing the individual patches instead of the entire breast image, our model enables precise detection of cancer tissues at a localized level.
Stats about breast cancer
According to the World Health Organization (WHO), in 2020 alone, there were 2.3 million reported cases of breast cancer among women, resulting in 685,000 deaths worldwide. By the end of the same year, there were approximately 7.8 million women who had been diagnosed with breast cancer within the past five years, establishing it as the most prevalent form of cancer globally.
Worldwide, female breast cancer is the fifth leading cause of death.
Introduction
As stated by Pamela Wright, the medical director of the Breast Center at Johns Hopkins, Invasive Ductal Carcinoma (IDC), also referred to as infiltrating ductal carcinoma, is the predominant type of breast cancer. It represents 80% of all breast cancer diagnoses. For further information about IDC, please refer to this article. In the context of this project, we have developed a classification model based on Convolutional Neural Networks (CNNs) using TensorFlow. The model utilizes Histopathology Images of patients to classify whether they have breast cancer or not!
Dataset
We are using the dataset from Kaggle. It can be accessed here. Following are some of the properties of this dataset:
The initial dataset comprised 162 slide images of breast cancer specimens scanned at a magnification of 40x. Due to their large dimensions, 277,524 patches measuring 50×50 pixels were extracted from these images to improve their manageability. These patches encompass the regions that contain Invasive Ductal Carcinoma (IDC), thereby enabling more efficient processing and analysis.
- 198,738 negative examples (i.e., no breast cancer)
- 78,786 positive examples (i.e., indicating breast cancer)
The dataset assigns a unique filename structure to each image, like:
u_xX_yY_classC.png
For example:
10253_idx5_x1351_y1101_class0.png
- "u" is patient id
- "u" is the patient ID (10253_idx5),
- "X" is the x-coordinate of where this patch was cropped from,
- "Y" is the y-coordinate of where this patch was cropped from, and
- "C" indicates the class where 0 is non-IDC and 1 is IDC.
Images
The following set of images are generated in this project.
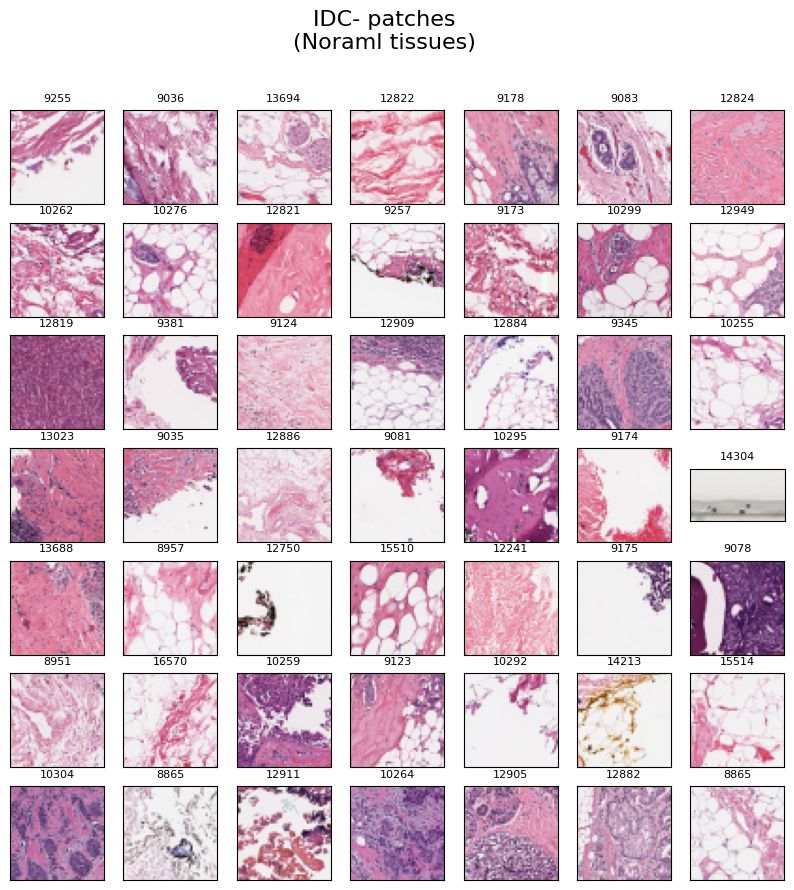
Normal Tissues
The image below displays a collection of 49 randomly selected IDC negative image patches, i.e., normal tissues. Each image is labeled with the respective "patient id" at the top.
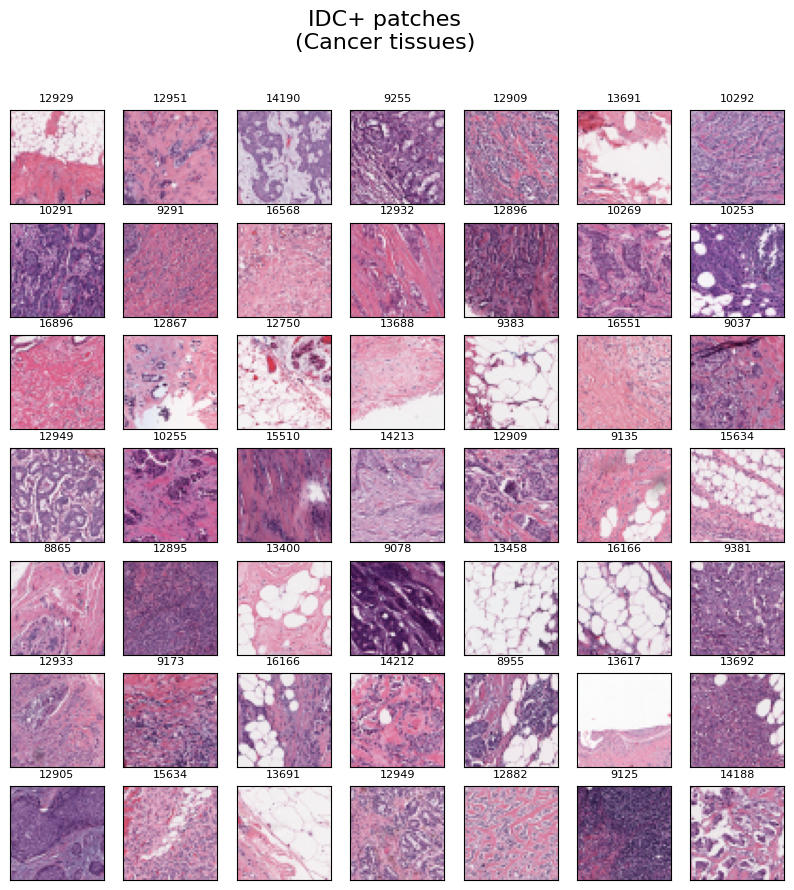
Cancer Tissues
Similarly, The provided image exhibits a set of 49 randomly chosen IDC positive image patches, which correspond to cancer tissues. Each image in the collection is accompanied by the corresponding "patient id" label positioned at the top.
Complete Histopathological image of breast
Presented below is the comprehensive Histopathological Image, revealing the entire breast tissue. This image has been formed by merging all the patches from the patient. Furthermore, a mask has been employed to accentuate the cancerous tissues, which are distinctly marked in green color.

Training hyperparameters
The following hyperparameters were used during training:
| Hyperparameters | Value |
|---|---|
| name | RMSprop |
| weight_decay | None |
| clipnorm | None |
| global_clipnorm | None |
| clipvalue | None |
| use_ema | False |
| ema_momentum | 0.99 |
| ema_overwrite_frequency | 100 |
| jit_compile | False |
| is_legacy_optimizer | False |
| learning_rate | 0.0010000000474974513 |
| rho | 0.9 |
| momentum | 0.0 |
| epsilon | 1e-07 |
| centered | False |
| training_precision | float32 |
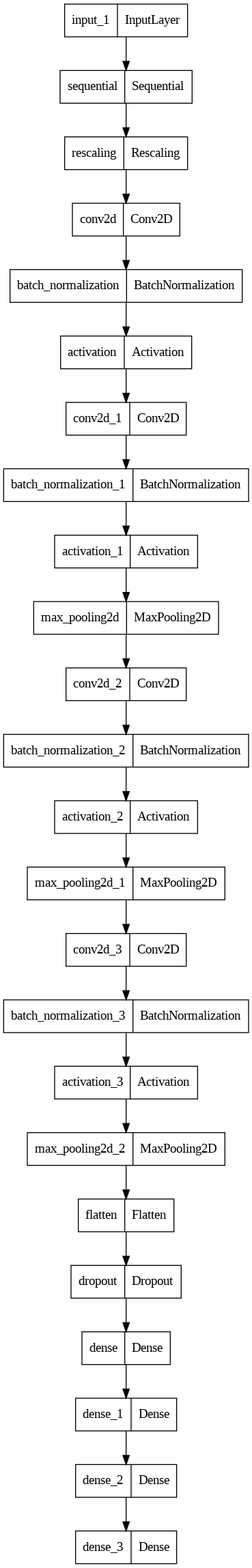
Model Plot
Conclusion
The test data evaluation of the model yielded exceptional results, with a test accuracy of 87%. This outcome is particularly impressive considering the small size of the dataset and the fact that the image patches were only 50x50 pixels, and the model was trained from scratch.
Furthermore, we discovered several key insights during the analysis:
In traditional Convolutional Neural Network (CNN) architectures, the number of filters typically increases progressively, following a pattern like 64, 128, and 256.
However, when we followed this conventional approach with our 50x50 input images, the accuracy on the test data was only 81%, and the validation graph showed significant fluctuations. The reason being that we lost neary half of the information after the first layer of CNN. Because the image dimention was reduced to 23x23.
By adopting a different filter configuration, specifically using 256 filters in each layer, we not only achieved a 6% increase in accuracy but also observed more stable fluctuations in the validation graph. This indicates that the model consistently generated accurate predictions.