Thiago
commited on
Commit
·
6b767f8
1
Parent(s):
f6b05c8
first commit
Browse files- README.md +29 -1
- app/.DS_Store +0 -0
- app/imgs/.DS_Store +0 -0
- app/imgs/app_1.png +0 -0
- app/imgs/app_2.png +0 -0
- app/imgs/doctor.png +0 -0
- app/imgs/emory_1.png +0 -0
- app/imgs/hybrid_system.png +0 -0
- app/imgs/icon.png +0 -0
- app/imgs/icons8-github-240.png +0 -0
- app/imgs/medical-checkup.png +0 -0
- app/imgs/pipeline.png +0 -0
- app/models/.DS_Store +0 -0
- app/models/all_labels_hierarchy/.gitignore +4 -0
- app/models/higher_order_hierarchy/.gitignore +4 -0
- app/src/__pycache__/app.cpython-38.pyc +0 -0
- app/src/__pycache__/config.cpython-37.pyc +0 -0
- app/src/__pycache__/config.cpython-38.pyc +0 -0
- app/src/__pycache__/download_models.cpython-37.pyc +0 -0
- app/src/__pycache__/pipeline.cpython-37.pyc +0 -0
- app/src/__pycache__/pipeline.cpython-38.pyc +0 -0
- app/src/__pycache__/pipeline.cpython-39.pyc +0 -0
- app/src/__pycache__/text_cleaning.cpython-37.pyc +0 -0
- app/src/__pycache__/text_cleaning.cpython-38.pyc +0 -0
- app/src/__pycache__/text_cleaning_transforerms.cpython-37.pyc +0 -0
- app/src/__pycache__/text_cleaning_transforerms.cpython-38.pyc +0 -0
- app/src/app.py +576 -0
- app/src/config.py +221 -0
- app/src/download_models.py +187 -0
- app/src/label_extraction.py +150 -0
- app/src/pipeline.py +670 -0
- app/src/text_cleaning.py +250 -0
- app/src/text_cleaning_transforerms.py +229 -0
- environment.yml +210 -0
- requirements.txt +180 -0
README.md
CHANGED
|
@@ -1 +1,29 @@
|
|
| 1 |
-
#
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Information Extraction from Breast Cancer Pathology Reports
|
| 2 |
+
|
| 3 |
+
<!-- TOC -->
|
| 4 |
+
|
| 5 |
+
- [Instruction Navigation](#information-extraction-from-breast-cancer-pathology-reports)
|
| 6 |
+
- [Web System Application](#web-system-application)
|
| 7 |
+
- [Model and Project Design](#model-and-project-design)
|
| 8 |
+
|
| 9 |
+
<!-- /TOC -->
|
| 10 |
+
|
| 11 |
+
|
| 12 |
+
## Web System Application
|
| 13 |
+
|
| 14 |
+
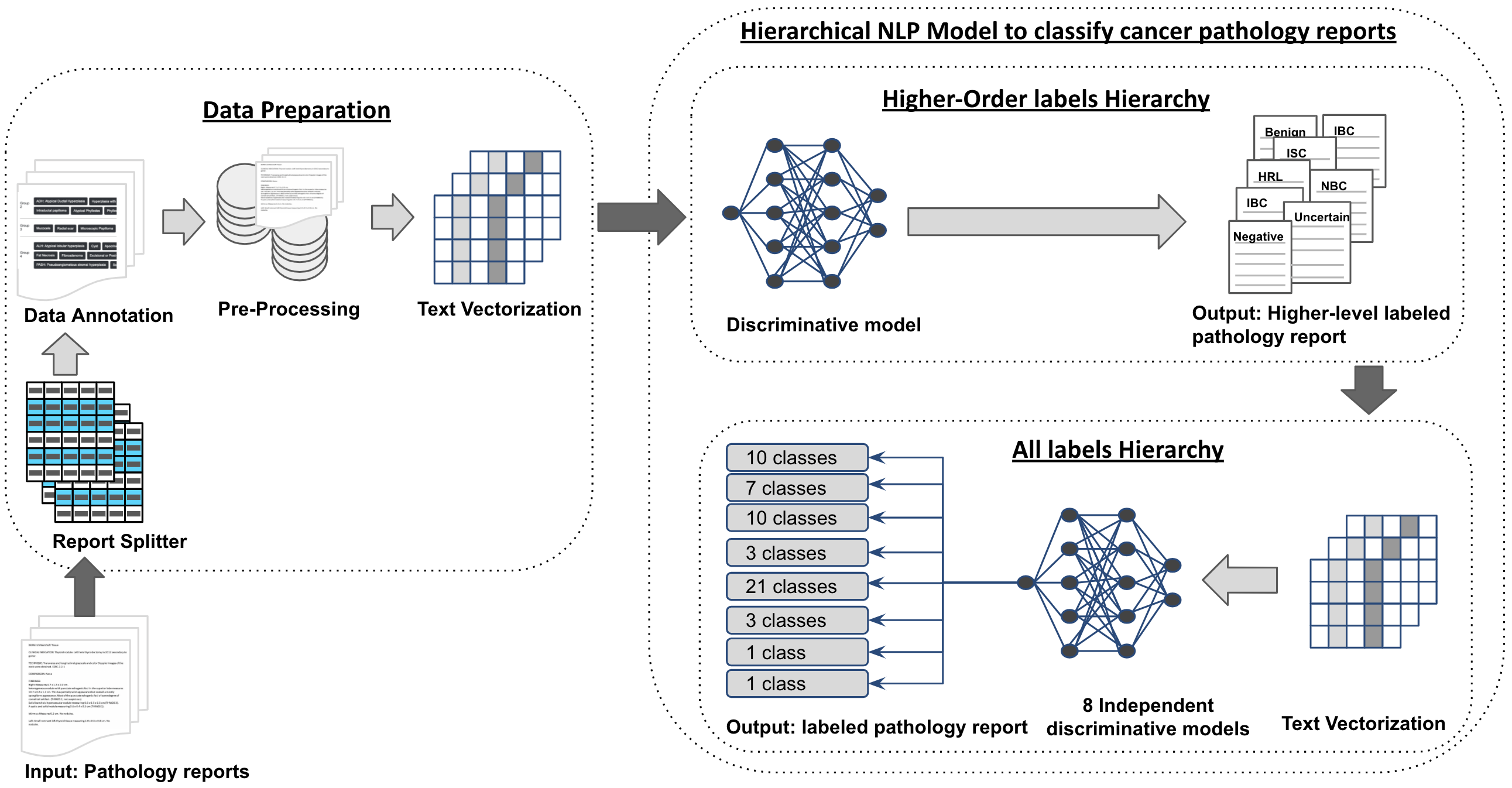
We developed a web system application for users to test our proposed pipilne for predicting histopathology reports. Users can interact with the platform in 2 ways: 1) Input an excel/csv spreadsheet with a column with the biopsy diagnosis (Part A,B or C). 2) Input a single biopsy diagnosis. An example of our Web System is illustraded bellow:
|
| 15 |
+
|
| 16 |
+
<table border=1>
|
| 17 |
+
<tr align='center' >
|
| 18 |
+
<td><img src="https://github.com/thiagosantos1/BreastPathologyClassificationSystem/blob/master/app/imgs/app_1.png" width="500" title="HCS App"></td>
|
| 19 |
+
<td><img src="https://github.com/thiagosantos1/BreastPathologyClassificationSystem/blob/master/app/imgs/app_2.png" width="500" title="HCS App"></td>
|
| 20 |
+
</tr>
|
| 21 |
+
</table>
|
| 22 |
+
|
| 23 |
+
## Model and Project Design
|
| 24 |
+
<table border=1>
|
| 25 |
+
<tr align='center' >
|
| 26 |
+
<td><img src="https://github.com/thiagosantos1/BreastPathologyClassificationSystem/blob/master/app/imgs/pipeline.png" width="500" title="HCS App"></td>
|
| 27 |
+
<td><img src="https://github.com/thiagosantos1/BreastPathologyClassificationSystem/blob/master/app/imgs/hybrid_system.png" width="400" title="HCS App"></td>
|
| 28 |
+
</tr>
|
| 29 |
+
</table>
|
app/.DS_Store
ADDED
|
Binary file (6.15 kB). View file
|
|
|
app/imgs/.DS_Store
ADDED
|
Binary file (6.15 kB). View file
|
|
|
app/imgs/app_1.png
ADDED

|
app/imgs/app_2.png
ADDED

|
app/imgs/doctor.png
ADDED

|
app/imgs/emory_1.png
ADDED

|
app/imgs/hybrid_system.png
ADDED

|
app/imgs/icon.png
ADDED
|
|
app/imgs/icons8-github-240.png
ADDED
|
|
app/imgs/medical-checkup.png
ADDED

|
app/imgs/pipeline.png
ADDED
|
app/models/.DS_Store
ADDED
|
Binary file (6.15 kB). View file
|
|
|
app/models/all_labels_hierarchy/.gitignore
ADDED
|
@@ -0,0 +1,4 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Ignore everything in this directory
|
| 2 |
+
*
|
| 3 |
+
# Except this file
|
| 4 |
+
!.gitignore
|
app/models/higher_order_hierarchy/.gitignore
ADDED
|
@@ -0,0 +1,4 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Ignore everything in this directory
|
| 2 |
+
*
|
| 3 |
+
# Except this file
|
| 4 |
+
!.gitignore
|
app/src/__pycache__/app.cpython-38.pyc
ADDED
|
Binary file (4.36 kB). View file
|
|
|
app/src/__pycache__/config.cpython-37.pyc
ADDED
|
Binary file (3.64 kB). View file
|
|
|
app/src/__pycache__/config.cpython-38.pyc
ADDED
|
Binary file (3.65 kB). View file
|
|
|
app/src/__pycache__/download_models.cpython-37.pyc
ADDED
|
Binary file (5.47 kB). View file
|
|
|
app/src/__pycache__/pipeline.cpython-37.pyc
ADDED
|
Binary file (20 kB). View file
|
|
|
app/src/__pycache__/pipeline.cpython-38.pyc
ADDED
|
Binary file (18.2 kB). View file
|
|
|
app/src/__pycache__/pipeline.cpython-39.pyc
ADDED
|
Binary file (11.7 kB). View file
|
|
|
app/src/__pycache__/text_cleaning.cpython-37.pyc
ADDED
|
Binary file (7.86 kB). View file
|
|
|
app/src/__pycache__/text_cleaning.cpython-38.pyc
ADDED
|
Binary file (8.01 kB). View file
|
|
|
app/src/__pycache__/text_cleaning_transforerms.cpython-37.pyc
ADDED
|
Binary file (6.09 kB). View file
|
|
|
app/src/__pycache__/text_cleaning_transforerms.cpython-38.pyc
ADDED
|
Binary file (6.5 kB). View file
|
|
|
app/src/app.py
ADDED
|
@@ -0,0 +1,576 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (C) 2021, Mindee.
|
| 2 |
+
|
| 3 |
+
# This program is licensed under the Apache License version 2.
|
| 4 |
+
# See LICENSE or go to <https://www.apache.org/licenses/LICENSE-2.0.txt> for full license details.
|
| 5 |
+
|
| 6 |
+
import os
|
| 7 |
+
import streamlit as st
|
| 8 |
+
import streamlit.components.v1 as components
|
| 9 |
+
import time
|
| 10 |
+
import matplotlib.pyplot as plt
|
| 11 |
+
import pandas as pd
|
| 12 |
+
from pipeline import Pipeline
|
| 13 |
+
import html
|
| 14 |
+
from IPython.core.display import display, HTML
|
| 15 |
+
import json
|
| 16 |
+
from PIL import Image
|
| 17 |
+
from tqdm import tqdm
|
| 18 |
+
import logging
|
| 19 |
+
from htbuilder import HtmlElement, div, ul, li, br, hr, a, p, img, styles, classes, fonts
|
| 20 |
+
from htbuilder.units import percent, px
|
| 21 |
+
from htbuilder.funcs import rgba, rgb
|
| 22 |
+
import copy
|
| 23 |
+
from download_models import check_if_exist
|
| 24 |
+
import re
|
| 25 |
+
import numpy as np
|
| 26 |
+
from sklearn.manifold import TSNE
|
| 27 |
+
from sklearn.decomposition import PCA
|
| 28 |
+
import plotly.express as plotpx
|
| 29 |
+
import umap
|
| 30 |
+
|
| 31 |
+
def image(src_as_string, **style):
|
| 32 |
+
return img(src=src_as_string, style=styles(**style))
|
| 33 |
+
|
| 34 |
+
|
| 35 |
+
def link(link, text, **style):
|
| 36 |
+
return a(_href=link, _target="_blank", style=styles(**style))(text)
|
| 37 |
+
|
| 38 |
+
def update_highlight(current,old):
|
| 39 |
+
out = current
|
| 40 |
+
matches_background_new = [(m.start(0), m.end(0)) for m in re.finditer("background-color:rgba\\(234, 131, 4,", out)]
|
| 41 |
+
matches_background_old = [(m.start(0), m.end(0)) for m in re.finditer("background-color:rgba\\(234, 131, 4,", old)]
|
| 42 |
+
for x,y in zip(matches_background_old,matches_background_new):
|
| 43 |
+
try:
|
| 44 |
+
old_importance = re.search("\\d+\\.\\d+",old[x[1]:x[1]+20])
|
| 45 |
+
new_importance = re.search("\\d+\\.\\d+",current[y[1]:y[1]+20])
|
| 46 |
+
|
| 47 |
+
if int(out[y[1]]) ==0 and float(old[x[1]]) != 0:
|
| 48 |
+
out = out[0:y[1]] + str(old_importance.group(0)) + out[y[1]:]
|
| 49 |
+
return False,out
|
| 50 |
+
if float(out[y[1]]) !=0 and float(old[x[1]]) != 0:
|
| 51 |
+
if float(old[x[1]]) > float(out[y[1]]):
|
| 52 |
+
out = out[0:y[1]] + str(old_importance.group(0))[0] + out[y[1]:]
|
| 53 |
+
return False,out
|
| 54 |
+
except Exception as e:
|
| 55 |
+
return True, out
|
| 56 |
+
|
| 57 |
+
return True,out
|
| 58 |
+
|
| 59 |
+
def hidde_menu():
|
| 60 |
+
|
| 61 |
+
footer_style = """<style>
|
| 62 |
+
footer {
|
| 63 |
+
visibility: hidden;
|
| 64 |
+
}
|
| 65 |
+
footer:after {
|
| 66 |
+
content:"An end-to-end Breast Pathology Classification System to infer Breast Cancer Diagnosis and Severity";
|
| 67 |
+
visibility: visible;
|
| 68 |
+
display: block;
|
| 69 |
+
position: center;
|
| 70 |
+
#background-color: red;
|
| 71 |
+
padding: 5px;
|
| 72 |
+
top: 2px;
|
| 73 |
+
}
|
| 74 |
+
</style>
|
| 75 |
+
"""
|
| 76 |
+
|
| 77 |
+
st.markdown(footer_style, unsafe_allow_html=True)
|
| 78 |
+
|
| 79 |
+
def main(myargs):
|
| 80 |
+
project_dir = os.path.dirname(os.path.abspath(__file__))
|
| 81 |
+
|
| 82 |
+
|
| 83 |
+
def add_content(columns):
|
| 84 |
+
if 'hg_df' in st.session_state:
|
| 85 |
+
columns[1].dataframe(st.session_state.hg_df)
|
| 86 |
+
if 'all_l' in st.session_state:
|
| 87 |
+
columns[2].dataframe(st.session_state.all_l)
|
| 88 |
+
|
| 89 |
+
if "highlight_samples" in st.session_state:
|
| 90 |
+
|
| 91 |
+
if "selected_indices" in st.session_state:
|
| 92 |
+
if len(st.session_state.selected_indices) >0:
|
| 93 |
+
out = ""
|
| 94 |
+
l = st.session_state.selected_indices
|
| 95 |
+
l.sort()
|
| 96 |
+
for ind in l:
|
| 97 |
+
out += st.session_state.highlight_samples[ind] + "<br><br>"
|
| 98 |
+
components.html(out,scrolling=True)
|
| 99 |
+
else:
|
| 100 |
+
components.html(st.session_state.highlight_samples[0])
|
| 101 |
+
else:
|
| 102 |
+
components.html(st.session_state.highlight_samples[0])
|
| 103 |
+
|
| 104 |
+
|
| 105 |
+
# Add Plot - Only for File version
|
| 106 |
+
if st.session_state['input_type'] == 'File' and "embeddings_all" in st.session_state and st.session_state.embeddings_plot in ["2D", "3D"]:
|
| 107 |
+
indices = [x for x in range(st.session_state.data_df[st.session_state.input_column].values.shape[0])]
|
| 108 |
+
if "selected_indices" in st.session_state:
|
| 109 |
+
if len(st.session_state.selected_indices) >=4:
|
| 110 |
+
l = st.session_state.selected_indices
|
| 111 |
+
l.sort()
|
| 112 |
+
indices = l
|
| 113 |
+
|
| 114 |
+
if st.session_state.data_df[st.session_state.input_column].values.shape[0] >=2:
|
| 115 |
+
sub_embeddings = st.session_state.embeddings_all[indices]
|
| 116 |
+
sentences = st.session_state.data_df[st.session_state.input_column].values[indices]
|
| 117 |
+
sentences_parses = []
|
| 118 |
+
break_size = 20
|
| 119 |
+
for data in sentences:
|
| 120 |
+
d = data.split()
|
| 121 |
+
size_sentence = len(d)
|
| 122 |
+
if len(d) >break_size:
|
| 123 |
+
out = ""
|
| 124 |
+
for lower_bound in range(0,size_sentence, break_size):
|
| 125 |
+
upper_bound = lower_bound + break_size if lower_bound + break_size <= size_sentence else size_sentence
|
| 126 |
+
out += " ".join(x for x in d[lower_bound:upper_bound]) + "<br>"
|
| 127 |
+
sentences_parses.append(out)
|
| 128 |
+
else:
|
| 129 |
+
sentences_parses.append(data)
|
| 130 |
+
|
| 131 |
+
|
| 132 |
+
|
| 133 |
+
prediction_label = st.session_state.hg_df["Prediction"].values[indices]
|
| 134 |
+
prediction_worst_label = []
|
| 135 |
+
for pred in prediction_label:
|
| 136 |
+
preds = pred.split(" && ")
|
| 137 |
+
if len(preds) ==1:
|
| 138 |
+
prediction_worst_label.extend(preds)
|
| 139 |
+
else:
|
| 140 |
+
worst_index = min([st.session_state.predictor.bert_model.config['worst_rank'].index(x) for x in preds])
|
| 141 |
+
prediction_worst_label.append(st.session_state.predictor.bert_model.config['worst_rank'][worst_index])
|
| 142 |
+
|
| 143 |
+
|
| 144 |
+
if st.session_state.embeddings_type == "PCA":
|
| 145 |
+
|
| 146 |
+
low_dim_embeddings = PCA(n_components=3).fit_transform(sub_embeddings)
|
| 147 |
+
elif st.session_state.embeddings_type == "TSNE":
|
| 148 |
+
low_dim_embeddings = TSNE(n_components=3,init="pca",perplexity=st.session_state.perplexity,learning_rate=st.session_state.learning_rate).fit_transform(sub_embeddings)
|
| 149 |
+
|
| 150 |
+
else:
|
| 151 |
+
n_neighbors = min(st.session_state.n_neighbors, len(sub_embeddings)-1 )
|
| 152 |
+
low_dim_embeddings = umap.UMAP(n_neighbors=n_neighbors, min_dist=st.session_state.min_dist,n_components=3).fit(sub_embeddings).embedding_
|
| 153 |
+
|
| 154 |
+
df_embeddings = pd.DataFrame(low_dim_embeddings)
|
| 155 |
+
df_embeddings = df_embeddings.rename(columns={0:'x',1:'y',2:'z'})
|
| 156 |
+
df_embeddings = df_embeddings.assign(severity=prediction_worst_label)
|
| 157 |
+
df_embeddings = df_embeddings.assign(text=sentences_parses)
|
| 158 |
+
df_embeddings = df_embeddings.assign(data_index=indices)
|
| 159 |
+
df_embeddings = df_embeddings.assign(all_predictions=prediction_label)
|
| 160 |
+
|
| 161 |
+
|
| 162 |
+
if st.session_state.embeddings_plot == "2D":
|
| 163 |
+
# 2D
|
| 164 |
+
plot = plotpx.scatter(
|
| 165 |
+
df_embeddings, x='x', y='y',
|
| 166 |
+
color='severity', labels={'color': 'severity'},
|
| 167 |
+
hover_data=['text','all_predictions','data_index'],title = 'BERT Embeddings Visualization - Please select rows (at least 4) to display specific examples'
|
| 168 |
+
)
|
| 169 |
+
|
| 170 |
+
else:
|
| 171 |
+
# 3D
|
| 172 |
+
plot = plotpx.scatter_3d(
|
| 173 |
+
df_embeddings, x='x', y='y', z='z',
|
| 174 |
+
color='severity', labels={'color': 'severity'},
|
| 175 |
+
hover_data=['text','all_predictions','data_index'],title = 'BERT Embeddings Visualization - Please select rows (at least 4) to display specific examples'
|
| 176 |
+
)
|
| 177 |
+
|
| 178 |
+
st.plotly_chart(plot,use_container_width=True,)
|
| 179 |
+
|
| 180 |
+
|
| 181 |
+
#worst_rank_ind = [classes.index(x) for x in worst_rank]
|
| 182 |
+
|
| 183 |
+
if 'bert_lime_output' in st.session_state and st.session_state.bert_lime:
|
| 184 |
+
if len(st.session_state.bert_lime_output) >0: # need to re-run prediction
|
| 185 |
+
st.markdown("BERT Interpretability")
|
| 186 |
+
components.html(st.session_state.bert_lime_output[0])
|
| 187 |
+
|
| 188 |
+
if 'json_output' in st.session_state and st.session_state.json_out:
|
| 189 |
+
|
| 190 |
+
st.markdown("Here are your analysis results in JSON format:")
|
| 191 |
+
out = {}
|
| 192 |
+
if "selected_indices" in st.session_state:
|
| 193 |
+
|
| 194 |
+
if len(st.session_state.selected_indices) >0:
|
| 195 |
+
l = st.session_state.selected_indices
|
| 196 |
+
l.sort()
|
| 197 |
+
for ind in l:
|
| 198 |
+
out['sample_'+str(ind)] = st.session_state.json_output['sample_'+str(ind)]
|
| 199 |
+
st.json(out)
|
| 200 |
+
else:
|
| 201 |
+
out['sample_'+str(0)] = st.session_state.json_output['sample_'+str(0)]
|
| 202 |
+
st.json(out)
|
| 203 |
+
else:
|
| 204 |
+
# Display JSON
|
| 205 |
+
out['sample_'+str(0)] = st.session_state.json_output['sample_'+str(0)]
|
| 206 |
+
st.json(out)
|
| 207 |
+
|
| 208 |
+
|
| 209 |
+
def delete_var_session(keys:list):
|
| 210 |
+
for key in keys:
|
| 211 |
+
if key in st.session_state:
|
| 212 |
+
del st.session_state[key]
|
| 213 |
+
|
| 214 |
+
im = Image.open(os.path.join(project_dir, "../imgs/icon.png"))
|
| 215 |
+
|
| 216 |
+
|
| 217 |
+
# Wide mode
|
| 218 |
+
st.set_page_config(page_title='HCSBC', layout = 'wide',page_icon=im,menu_items={
|
| 219 |
+
'Get Help': 'https://github.com/thiagosantos1/BreastPathologyClassificationSystem',
|
| 220 |
+
'Report a bug': "https://github.com/thiagosantos1/BreastPathologyClassificationSystem",
|
| 221 |
+
'About': "An end-to-end breast pathology classification system https://github.com/thiagosantos1/BreastPathologyClassificationSystem"
|
| 222 |
+
})
|
| 223 |
+
st.sidebar.image(os.path.join(project_dir,"../imgs/doctor.png"),use_column_width=False)
|
| 224 |
+
|
| 225 |
+
# Designing the interface
|
| 226 |
+
st.markdown("<h1 style='text-align: center; color: black;'>HCSBC: Hierarchical Classification System for Breast Cancer</h1>", unsafe_allow_html=True)
|
| 227 |
+
st.markdown("System Pipeline: Pathology Emory Pubmed BERT + 6 independent Machine Learning discriminators")
|
| 228 |
+
# For newline
|
| 229 |
+
st.write('\n')
|
| 230 |
+
# Instructions
|
| 231 |
+
st.markdown("*Hint: click on the top-right corner to enlarge it!*")
|
| 232 |
+
# Set the columns
|
| 233 |
+
|
| 234 |
+
cols = st.columns((1, 1, 1))
|
| 235 |
+
#cols = st.columns(4)
|
| 236 |
+
cols[0].subheader("Input Data")
|
| 237 |
+
cols[1].subheader("Severity Predictions")
|
| 238 |
+
cols[2].subheader("Diagnose Predictions")
|
| 239 |
+
|
| 240 |
+
# Sidebar
|
| 241 |
+
# File selection
|
| 242 |
+
st.sidebar.title("Data Selection")
|
| 243 |
+
|
| 244 |
+
st.session_state['input_type'] = st.sidebar.radio("Input Selection", ('File', 'Text'), key="data_format")
|
| 245 |
+
if "prev_input_type" not in st.session_state:
|
| 246 |
+
st.session_state['prev_input_type'] = st.session_state.input_type
|
| 247 |
+
|
| 248 |
+
st.write('<style>div.row-widget.stRadio > div{flex-direction:row;}</style>', unsafe_allow_html=True)
|
| 249 |
+
|
| 250 |
+
|
| 251 |
+
# Disabling warning
|
| 252 |
+
st.set_option('deprecation.showfileUploaderEncoding', False)
|
| 253 |
+
|
| 254 |
+
|
| 255 |
+
if st.session_state['input_type'] == 'File':
|
| 256 |
+
if st.session_state['prev_input_type'] == 'Text':
|
| 257 |
+
delete_var_session(keys=["data_df","data_columns","hg_df","all_l","highlight_samples","selected_indices","json_output","bert_lime_output","embeddings_all"])
|
| 258 |
+
st.session_state['prev_input_type'] = "File"
|
| 259 |
+
|
| 260 |
+
# Choose your own file
|
| 261 |
+
new_file = st.sidebar.file_uploader("Upload Document", type=['xlsx','csv'])
|
| 262 |
+
if 'uploaded_file' in st.session_state and st.session_state.uploaded_file != None and new_file != None:
|
| 263 |
+
if st.session_state.uploaded_file.name != new_file.name and st.session_state.uploaded_file.id != new_file.id:
|
| 264 |
+
delete_var_session(keys=["data_df","data_columns","hg_df","all_l","highlight_samples","selected_indices","json_output","bert_lime_output","embeddings_all"])
|
| 265 |
+
|
| 266 |
+
st.session_state['uploaded_file'] = new_file
|
| 267 |
+
|
| 268 |
+
data_columns = ['Input']
|
| 269 |
+
if 'data_columns' not in st.session_state:
|
| 270 |
+
st.session_state['data_columns'] = data_columns
|
| 271 |
+
|
| 272 |
+
if st.session_state.uploaded_file is not None:
|
| 273 |
+
if 'data_df' not in st.session_state:
|
| 274 |
+
if st.session_state.uploaded_file.name.endswith('.xlsx'):
|
| 275 |
+
df = pd.read_excel(st.session_state.uploaded_file)
|
| 276 |
+
else:
|
| 277 |
+
df = pd.read_csv(st.session_state.uploaded_file)
|
| 278 |
+
|
| 279 |
+
df = df.loc[:, ~df.columns.str.contains('^Unnamed')]
|
| 280 |
+
df = df.fillna("NA")
|
| 281 |
+
data_columns = df.columns.values
|
| 282 |
+
st.session_state['data_df'] = df
|
| 283 |
+
st.session_state['data_columns'] = data_columns
|
| 284 |
+
else:
|
| 285 |
+
if st.session_state['prev_input_type'] == 'File':
|
| 286 |
+
delete_var_session(keys=["data_df","input_column","user_input","hg_df","all_l","highlight_samples","selected_indices","json_output","bert_lime_output","embeddings_all"])
|
| 287 |
+
st.session_state['prev_input_type'] = "Text"
|
| 288 |
+
|
| 289 |
+
input_column = "Input"
|
| 290 |
+
data = st.sidebar.text_area("Please enter a breast cancer pathology diagnose")
|
| 291 |
+
if "user_input" in st.session_state:
|
| 292 |
+
if data != st.session_state.user_input:
|
| 293 |
+
delete_var_session(keys=["data_df","input_column","user_input","hg_df","all_l","highlight_samples","selected_indices","json_output","bert_lime_output","embeddings_all"])
|
| 294 |
+
|
| 295 |
+
st.session_state['user_input'] = data
|
| 296 |
+
if len(st.session_state.user_input.split()) >0:
|
| 297 |
+
st.session_state['data_df'] = pd.DataFrame([st.session_state['user_input']], columns =[input_column])
|
| 298 |
+
st.session_state['input_column'] = input_column
|
| 299 |
+
st.session_state['uploaded_file'] = True
|
| 300 |
+
else:
|
| 301 |
+
delete_var_session(keys=["data_df","input_column","user_input","hg_df","all_l","highlight_samples","selected_indices","json_output","bert_lime_output","embeddings_all"])
|
| 302 |
+
|
| 303 |
+
|
| 304 |
+
if 'data_df' in st.session_state:
|
| 305 |
+
cols[0].dataframe(st.session_state.data_df)
|
| 306 |
+
|
| 307 |
+
|
| 308 |
+
if st.session_state['input_type'] == 'File':
|
| 309 |
+
# Columns selection
|
| 310 |
+
st.sidebar.write('\n')
|
| 311 |
+
st.sidebar.title("Column For Prediction")
|
| 312 |
+
input_column = st.sidebar.selectbox("Columns", st.session_state.data_columns)
|
| 313 |
+
|
| 314 |
+
st.session_state['input_column'] = input_column
|
| 315 |
+
|
| 316 |
+
|
| 317 |
+
st.sidebar.write('\n')
|
| 318 |
+
st.sidebar.title("Severity Model")
|
| 319 |
+
input_higher = st.sidebar.selectbox("Model", ["PathologyEmoryPubMedBERT"])
|
| 320 |
+
st.session_state['input_higher'] = input_higher
|
| 321 |
+
|
| 322 |
+
if "prev_input_higher" not in st.session_state:
|
| 323 |
+
st.session_state['prev_input_higher'] = st.session_state.input_higher
|
| 324 |
+
st.session_state['input_higher_exist'] = check_if_exist(st.session_state.input_higher)
|
| 325 |
+
st.session_state['load_new_higher_model'] = True
|
| 326 |
+
elif st.session_state.prev_input_higher != st.session_state.input_higher:
|
| 327 |
+
st.session_state['input_higher_exist'] = check_if_exist(st.session_state.input_higher)
|
| 328 |
+
st.session_state['prev_input_higher'] = st.session_state.input_higher
|
| 329 |
+
st.session_state['load_new_higher_model'] = True
|
| 330 |
+
delete_var_session(keys=["data_df","input_column","user_input","hg_df","all_l","highlight_samples","selected_indices","json_output","bert_lime_output","embeddings_all"])
|
| 331 |
+
|
| 332 |
+
|
| 333 |
+
st.sidebar.write('\n')
|
| 334 |
+
st.sidebar.title("Diagnosis Model")
|
| 335 |
+
input_all_labels = st.sidebar.selectbox("Model", ['single_vectorizer', 'branch_vectorizer'])
|
| 336 |
+
st.session_state['input_all_labels'] = input_all_labels
|
| 337 |
+
|
| 338 |
+
if "prev_input_all_labels" not in st.session_state:
|
| 339 |
+
st.session_state['prev_input_all_labels'] = st.session_state.input_all_labels
|
| 340 |
+
st.session_state['input_all_labels_exist'] = check_if_exist(st.session_state.input_all_labels)
|
| 341 |
+
st.session_state['load_new_all_label_model'] = True
|
| 342 |
+
elif st.session_state.prev_input_all_labels != st.session_state.input_all_labels:
|
| 343 |
+
st.session_state['input_all_labels_exist'] = check_if_exist(st.session_state.input_all_labels)
|
| 344 |
+
st.session_state['prev_input_all_labels'] = st.session_state.input_all_labels
|
| 345 |
+
st.session_state['load_new_all_label_model'] = True
|
| 346 |
+
delete_var_session(keys=["data_df","input_column","user_input","hg_df","all_l","highlight_samples","selected_indices","json_output","bert_lime_output","embeddings_all"])
|
| 347 |
+
|
| 348 |
+
|
| 349 |
+
# For newline
|
| 350 |
+
st.sidebar.write('\n')
|
| 351 |
+
st.sidebar.title("Analysis Options")
|
| 352 |
+
|
| 353 |
+
predictions, json_output, higher_order_pred,all_labels_pred,higher_order_prob,all_labels_prob = {},[],[],[],[],[]
|
| 354 |
+
hg_df, all_l,highlight_samples, bert_lime_output, embeddings_all= [],[],[],[],[]
|
| 355 |
+
|
| 356 |
+
|
| 357 |
+
if st.session_state['input_type'] == 'File':
|
| 358 |
+
embeddings_plot = st.sidebar.radio('Display embeddings plot',
|
| 359 |
+
['2D',
|
| 360 |
+
'3D',
|
| 361 |
+
'Dont Display'],index=1)
|
| 362 |
+
|
| 363 |
+
st.session_state['embeddings_plot'] = embeddings_plot
|
| 364 |
+
|
| 365 |
+
else:
|
| 366 |
+
st.session_state['embeddings_plot'] = 'Dont Display'
|
| 367 |
+
|
| 368 |
+
if st.session_state['input_type'] == 'File':
|
| 369 |
+
embeddings_type = st.sidebar.radio('Dimensionality Reduction',
|
| 370 |
+
['PCA',
|
| 371 |
+
'TSNE','UMAP'],index=0)
|
| 372 |
+
|
| 373 |
+
st.session_state['embeddings_type'] = embeddings_type
|
| 374 |
+
|
| 375 |
+
if st.session_state.embeddings_type == "TSNE":
|
| 376 |
+
perplexity = st.sidebar.slider("Perplexity", min_value=5, max_value=100, step=5, value=30)
|
| 377 |
+
st.session_state['perplexity'] = perplexity
|
| 378 |
+
|
| 379 |
+
learning_rate = st.sidebar.slider("Learning Rate", min_value=10, max_value=1000, step=10, value=100)
|
| 380 |
+
st.session_state['learning_rate'] = learning_rate
|
| 381 |
+
|
| 382 |
+
if st.session_state.embeddings_type == "UMAP":
|
| 383 |
+
n_neighbors = st.sidebar.slider("Neighbors", min_value=2, max_value=100, step=1, value=2)
|
| 384 |
+
st.session_state['n_neighbors'] = n_neighbors
|
| 385 |
+
|
| 386 |
+
min_dist = st.sidebar.slider("Minimal Distance", min_value=0.1, max_value=0.99, step=0.05, value=0.1)
|
| 387 |
+
st.session_state['min_dist'] = min_dist
|
| 388 |
+
|
| 389 |
+
json_out = st.sidebar.checkbox('Display Json',value = True,key='check3')
|
| 390 |
+
st.session_state['json_out'] = json_out
|
| 391 |
+
|
| 392 |
+
if st.session_state['input_type'] == 'Text':
|
| 393 |
+
bert_lime = st.sidebar.checkbox('Display BERT Interpretability',value = False,key='check3')
|
| 394 |
+
st.session_state['bert_lime'] = bert_lime
|
| 395 |
+
else:
|
| 396 |
+
st.session_state['bert_lime'] = False
|
| 397 |
+
|
| 398 |
+
|
| 399 |
+
# For newline
|
| 400 |
+
st.sidebar.write('\n')
|
| 401 |
+
st.sidebar.title("Prediction")
|
| 402 |
+
|
| 403 |
+
|
| 404 |
+
if st.sidebar.button("Run Prediction"):
|
| 405 |
+
|
| 406 |
+
if st.session_state.uploaded_file is None:
|
| 407 |
+
st.sidebar.write("Please upload a your data")
|
| 408 |
+
|
| 409 |
+
else:
|
| 410 |
+
st.session_state['input_all_labels_exist'] = check_if_exist(st.session_state.input_all_labels)
|
| 411 |
+
if not st.session_state.input_all_labels_exist:
|
| 412 |
+
st.sidebar.write("Please Download Model: " + str(st.session_state.input_all_labels))
|
| 413 |
+
|
| 414 |
+
st.session_state['input_higher_exist'] = check_if_exist(st.session_state.input_higher)
|
| 415 |
+
if not st.session_state.input_higher_exist:
|
| 416 |
+
st.sidebar.write("Please Download Model: " + str(st.session_state.input_higher))
|
| 417 |
+
|
| 418 |
+
if st.session_state.input_all_labels_exist and st.session_state.input_higher_exist:
|
| 419 |
+
if "predictor" not in st.session_state or st.session_state.load_new_higher_model or st.session_state.load_new_all_label_model:
|
| 420 |
+
with st.spinner('Loading model...'):
|
| 421 |
+
print("\n\tLoading Model")
|
| 422 |
+
st.session_state["predictor"] = Pipeline(bert_option=str(st.session_state.input_higher), branch_option=str(st.session_state.input_all_labels))
|
| 423 |
+
st.session_state['load_new_higher_model'] = False
|
| 424 |
+
st.session_state['load_new_all_label_model'] = False
|
| 425 |
+
|
| 426 |
+
with st.spinner('Transforming Data...'):
|
| 427 |
+
data = st.session_state.data_df[st.session_state.input_column].values
|
| 428 |
+
|
| 429 |
+
with st.spinner('Analyzing...'):
|
| 430 |
+
time.sleep(0.1)
|
| 431 |
+
prog_bar = st.progress(0)
|
| 432 |
+
logging.info("Running Predictions for data size of: " + str(len(data)))
|
| 433 |
+
logging.info("\n\tRunning Predictions with: " + str(st.session_state.input_higher) + str(st.session_state.input_all_labels))
|
| 434 |
+
for index in tqdm(range(len(data))):
|
| 435 |
+
d = data[index]
|
| 436 |
+
time.sleep(0.1)
|
| 437 |
+
prog_bar.progress(int( (100/len(data)) * (index+1) ))
|
| 438 |
+
# refactor json
|
| 439 |
+
preds,embeddings_output = st.session_state.predictor.run(d)
|
| 440 |
+
embeddings = embeddings_output.tolist()
|
| 441 |
+
embeddings_all.append(embeddings[0])
|
| 442 |
+
if st.session_state.bert_lime:
|
| 443 |
+
logging.info("Running BERT LIME Interpretability Predictions")
|
| 444 |
+
bert_lime_output.append(st.session_state.predictor.bert_interpretability(d))
|
| 445 |
+
|
| 446 |
+
predictions["sample_" + str(index)] = {}
|
| 447 |
+
for ind,pred in enumerate(preds):
|
| 448 |
+
predictions["sample_" + str(index)]["prediction_" + str(ind)] = pred
|
| 449 |
+
|
| 450 |
+
|
| 451 |
+
prog_bar.progress(100)
|
| 452 |
+
time.sleep(0.1)
|
| 453 |
+
|
| 454 |
+
for key,sample in predictions.items():
|
| 455 |
+
higher,all_p, prob_higher, prob_all = [],[],[],[]
|
| 456 |
+
for key,pred in sample.items():
|
| 457 |
+
for higher_order, sub_arr in pred.items():
|
| 458 |
+
higher.append(higher_order)
|
| 459 |
+
prob_higher.append(round(sub_arr["probability"], 2))
|
| 460 |
+
for label,v in sub_arr['labels'].items():
|
| 461 |
+
all_p.append(label)
|
| 462 |
+
prob_all.append(round(v["probability"], 2))
|
| 463 |
+
|
| 464 |
+
higher_order_pred.append(" && ".join(x for x in higher))
|
| 465 |
+
all_labels_pred.append(" && ".join(x for x in all_p))
|
| 466 |
+
|
| 467 |
+
higher_order_prob.append(" && ".join(str(x) for x in prob_higher))
|
| 468 |
+
all_labels_prob.append(" && ".join(str(x) for x in prob_all))
|
| 469 |
+
|
| 470 |
+
predictions_refact = copy.deepcopy(predictions)
|
| 471 |
+
|
| 472 |
+
for index in tqdm(range(len(data))):
|
| 473 |
+
highlights = ""
|
| 474 |
+
key = "sample_" + str(index)
|
| 475 |
+
for k,v in predictions[key].items():
|
| 476 |
+
for k_s, v_s in v.items():
|
| 477 |
+
predictions_refact["sample_" + str(index)]["data"] = v_s['data']
|
| 478 |
+
predictions_refact["sample_" + str(index)]["transformer_data"] = v_s['transformer_data']
|
| 479 |
+
predictions_refact["sample_" + str(index)]["discriminator_data"] = v_s['word_analysis']['discriminator_data']
|
| 480 |
+
highlight = v_s['word_analysis']['highlighted_html_text']
|
| 481 |
+
|
| 482 |
+
if len(highlights) >0:
|
| 483 |
+
done = False
|
| 484 |
+
merged = highlight
|
| 485 |
+
while not done:
|
| 486 |
+
done,merged = update_highlight(merged,highlights)
|
| 487 |
+
|
| 488 |
+
highlights = merged
|
| 489 |
+
else:
|
| 490 |
+
highlights = highlight
|
| 491 |
+
|
| 492 |
+
del predictions_refact[key][k][k_s]['data']
|
| 493 |
+
del predictions_refact[key][k][k_s]['transformer_data']
|
| 494 |
+
del predictions_refact[key][k][k_s]['word_analysis']['discriminator_data']
|
| 495 |
+
|
| 496 |
+
highlight_samples.append(highlights)
|
| 497 |
+
|
| 498 |
+
json_output = predictions_refact
|
| 499 |
+
|
| 500 |
+
hg_df = pd.DataFrame(list(zip(higher_order_pred, higher_order_prob)), columns =['Prediction', "Probability"])
|
| 501 |
+
all_l = pd.DataFrame(list(zip(all_labels_pred,all_labels_prob)), columns =['Prediction',"Probability"])
|
| 502 |
+
all_preds = pd.DataFrame(list(zip(higher_order_pred, all_labels_pred)), columns =['Severity Prediction',"Diagnose Prediction"])
|
| 503 |
+
|
| 504 |
+
st.session_state['hg_df'] = hg_df
|
| 505 |
+
st.session_state['all_l'] = all_l
|
| 506 |
+
st.session_state['all_preds'] = all_preds
|
| 507 |
+
st.session_state['json_output'] = json_output
|
| 508 |
+
st.session_state['highlight_samples'] = highlight_samples
|
| 509 |
+
st.session_state['highlight_samples_df'] = pd.DataFrame(highlight_samples, columns =["HTML Word Importance"])
|
| 510 |
+
st.session_state['bert_lime_output'] = bert_lime_output
|
| 511 |
+
st.session_state['embeddings_all'] = np.asarray(embeddings_all)
|
| 512 |
+
|
| 513 |
+
if 'data_df' in st.session_state and 'json_output' in st.session_state:
|
| 514 |
+
st.markdown("<h1 style='text-align: center; color: purple;'>Model Analysis</h1>", unsafe_allow_html=True)
|
| 515 |
+
selected_indices = st.multiselect('Select Rows to Display Word Importance, Embeddings Visualization, and Json Analysis:', [x for x in range(len(st.session_state.data_df))])
|
| 516 |
+
st.session_state['selected_indices'] = selected_indices
|
| 517 |
+
|
| 518 |
+
add_content(cols)
|
| 519 |
+
|
| 520 |
+
|
| 521 |
+
if 'json_output' in st.session_state:
|
| 522 |
+
st.sidebar.write('\n')
|
| 523 |
+
st.sidebar.title("Save Results")
|
| 524 |
+
|
| 525 |
+
st.sidebar.write('\n')
|
| 526 |
+
st.sidebar.download_button(
|
| 527 |
+
label="Download Output Json",
|
| 528 |
+
data=str(st.session_state.json_output),
|
| 529 |
+
file_name="output.json",
|
| 530 |
+
)
|
| 531 |
+
st.sidebar.download_button(
|
| 532 |
+
label="Download Predictions",
|
| 533 |
+
data=st.session_state.all_preds.to_csv(),
|
| 534 |
+
file_name="predictions.csv",
|
| 535 |
+
)
|
| 536 |
+
st.sidebar.download_button(
|
| 537 |
+
label="Download Data + Predictions",
|
| 538 |
+
data = pd.concat([st.session_state.data_df, st.session_state.all_preds,st.session_state.highlight_samples_df], axis=1, join='inner').to_csv(),
|
| 539 |
+
file_name="data_predictions.csv",
|
| 540 |
+
)
|
| 541 |
+
|
| 542 |
+
st.sidebar.write('\n')
|
| 543 |
+
st.sidebar.title("Contact Me")
|
| 544 |
+
sub_colms = st.sidebar.columns([1, 1, 1])
|
| 545 |
+
sub_colms[0].markdown('''<a href="https://github.com/thiagosantos1/BreastPathologyClassificationSystem">
|
| 546 |
+
<img src="https://img.icons8.com/fluency/48/000000/github.png" /></a>''',unsafe_allow_html=True)
|
| 547 |
+
sub_colms[1].markdown('''<a href="https://twitter.com/intent/follow?original_referer=https%3A%2F%2Fgithub.com%2Ftsantos_maia&screen_name=tsantos_maia">
|
| 548 |
+
<img src="https://img.icons8.com/color/48/000000/twitter--v1.png" /></a>''',unsafe_allow_html=True)
|
| 549 |
+
sub_colms[2].markdown('''<a href="https://www.linkedin.com/in/thiagosantos-cs/">
|
| 550 |
+
<img src="https://img.icons8.com/color/48/000000/linkedin.png" /></a>''',unsafe_allow_html=True)
|
| 551 |
+
|
| 552 |
+
|
| 553 |
+
hidde_menu()
|
| 554 |
+
|
| 555 |
+
|
| 556 |
+
|
| 557 |
+
|
| 558 |
+
if __name__ == '__main__':
|
| 559 |
+
|
| 560 |
+
myargs = [
|
| 561 |
+
"Made in ",
|
| 562 |
+
image('https://avatars3.githubusercontent.com/u/45109972?s=400&v=4',
|
| 563 |
+
width=px(25), height=px(25)),
|
| 564 |
+
" with ❤️ by ",
|
| 565 |
+
link("https://www.linkedin.com/in/thiagosantos-cs/", "@thiagosantos-cs"),
|
| 566 |
+
br(),
|
| 567 |
+
link("https://www.linkedin.com/in/thiagosantos-cs/", image('https://img.icons8.com/color/48/000000/twitter--v1.png')),
|
| 568 |
+
link("https://github.com/thiagosantos1/BreastPathologyClassificationSystem", image('https://img.icons8.com/fluency/48/000000/github.png')),
|
| 569 |
+
]
|
| 570 |
+
logging.basicConfig(
|
| 571 |
+
format="%(asctime)s - %(levelname)s - %(filename)s - %(message)s",
|
| 572 |
+
datefmt="%d/%m/%Y %H:%M:%S",
|
| 573 |
+
level=logging.INFO)
|
| 574 |
+
main(myargs)
|
| 575 |
+
|
| 576 |
+
|
app/src/config.py
ADDED
|
@@ -0,0 +1,221 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""
|
| 2 |
+
Input config for pipeline
|
| 3 |
+
"""
|
| 4 |
+
|
| 5 |
+
def config_file() -> dict:
|
| 6 |
+
config = {
|
| 7 |
+
"BERT_config": {
|
| 8 |
+
"model_emb": 'bert',
|
| 9 |
+
|
| 10 |
+
"model_option": {
|
| 11 |
+
"PathologyEmoryPubMedBERT": {
|
| 12 |
+
"model_folder":"../models/higher_order_hierarchy/PathologyEmoryPubMedBERT/"
|
| 13 |
+
},
|
| 14 |
+
"PathologyEmoryBERT": {
|
| 15 |
+
"model_folder":"../models/higher_order_hierarchy/PathologyEmoryBERT/"
|
| 16 |
+
},
|
| 17 |
+
"ClinicalBERT": {
|
| 18 |
+
"model_folder":"../models/higher_order_hierarchy/ClinicalBERT/"
|
| 19 |
+
},
|
| 20 |
+
"BlueBERT": {
|
| 21 |
+
"model_folder":"../models/higher_order_hierarchy/BlueBERT/"
|
| 22 |
+
},
|
| 23 |
+
"BioBERT": {
|
| 24 |
+
"model_folder":"../models/higher_order_hierarchy/BioBERT/"
|
| 25 |
+
},
|
| 26 |
+
"BERT": {
|
| 27 |
+
"model_folder":"../models/higher_order_hierarchy/BERT/"
|
| 28 |
+
},
|
| 29 |
+
|
| 30 |
+
},
|
| 31 |
+
"max_seq_length": "64",
|
| 32 |
+
"threshold_prediction":0.5,
|
| 33 |
+
"classes": ['Invasive breast cancer-IBC','Non-breast cancer-NBC','In situ breast cancer-ISC',
|
| 34 |
+
'Borderline lesion-BLL','High risk lesion-HRL','Benign-B','Negative'],
|
| 35 |
+
"worst_rank" : ['Invasive breast cancer-IBC', 'In situ breast cancer-ISC', 'High risk lesion-HRL',
|
| 36 |
+
'Borderline lesion-BLL','Benign-B','Non-breast cancer-NBC','Negative']
|
| 37 |
+
},
|
| 38 |
+
|
| 39 |
+
|
| 40 |
+
"ibc_config": {
|
| 41 |
+
|
| 42 |
+
"model_option": {
|
| 43 |
+
"single_tfidf": {
|
| 44 |
+
"path_model":"../models/all_labels_hierarchy/single_tfidf/classifiers",
|
| 45 |
+
"model": "ibc_xgboost_classifier.pkl",
|
| 46 |
+
"path_vectorizer":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 47 |
+
"vectorizer":"vectorizer_all_branches.pkl",
|
| 48 |
+
"path_bigrmas":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 49 |
+
"bigrams":"best_bigrams.csv",
|
| 50 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 51 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 52 |
+
},
|
| 53 |
+
|
| 54 |
+
"branch_tfidf": {
|
| 55 |
+
"path_model":"../models/all_labels_hierarchy/branch_tfidf/classifiers",
|
| 56 |
+
"model": "ibc_xgboost_classifier.pkl",
|
| 57 |
+
"path_vectorizer":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 58 |
+
"vectorizer":"ibc_vectorizer.pkl",
|
| 59 |
+
"path_bigrmas":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 60 |
+
"bigrams":"best_bigrams.csv",
|
| 61 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 62 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 63 |
+
}
|
| 64 |
+
},
|
| 65 |
+
|
| 66 |
+
"classes": ['apocrine carcinoma','grade i','grade ii','grade iii','invasive ductal carcinoma','invasive lobular carcinoma','medullary carcinoma','metaplastic carcinoma','mucinous carcinoma','tubular carcinoma','lymph node - metastatic']
|
| 67 |
+
|
| 68 |
+
},
|
| 69 |
+
|
| 70 |
+
"isc_config": {
|
| 71 |
+
"model_option": {
|
| 72 |
+
"single_tfidf": {
|
| 73 |
+
"path_model":"../models/all_labels_hierarchy/single_tfidf/classifiers",
|
| 74 |
+
"model": "isc_xgboost_classifier.pkl",
|
| 75 |
+
"path_vectorizer":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 76 |
+
"vectorizer":"vectorizer_all_branches.pkl",
|
| 77 |
+
"path_bigrmas":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 78 |
+
"bigrams":"best_bigrams.csv",
|
| 79 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 80 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 81 |
+
},
|
| 82 |
+
|
| 83 |
+
"branch_tfidf": {
|
| 84 |
+
"path_model":"../models/all_labels_hierarchy/branch_tfidf/classifiers",
|
| 85 |
+
"model": "isc_xgboost_classifier.pkl",
|
| 86 |
+
"path_vectorizer":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 87 |
+
"vectorizer":"isc_vectorizer.pkl",
|
| 88 |
+
"path_bigrmas":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 89 |
+
"bigrams":"best_bigrams.csv",
|
| 90 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 91 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 92 |
+
}
|
| 93 |
+
},
|
| 94 |
+
|
| 95 |
+
|
| 96 |
+
"classes": ['ductal carcinoma in situ','high','intermediate','intracystic papillary carcinoma','intraductal papillary carcinoma','low','pagets','fna - malignant']
|
| 97 |
+
|
| 98 |
+
},
|
| 99 |
+
|
| 100 |
+
"hrl_config": {
|
| 101 |
+
"model_option": {
|
| 102 |
+
"single_tfidf": {
|
| 103 |
+
"path_model":"../models/all_labels_hierarchy/single_tfidf/classifiers",
|
| 104 |
+
"model": "hrl_xgboost_classifier.pkl",
|
| 105 |
+
"path_vectorizer":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 106 |
+
"vectorizer":"vectorizer_all_branches.pkl",
|
| 107 |
+
"path_bigrmas":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 108 |
+
"bigrams":"best_bigrams.csv",
|
| 109 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 110 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 111 |
+
},
|
| 112 |
+
|
| 113 |
+
"branch_tfidf": {
|
| 114 |
+
"path_model":"../models/all_labels_hierarchy/branch_tfidf/classifiers",
|
| 115 |
+
"model": "hrl_xgboost_classifier.pkl",
|
| 116 |
+
"path_vectorizer":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 117 |
+
"vectorizer":"hrl_vectorizer.pkl",
|
| 118 |
+
"path_bigrmas":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 119 |
+
"bigrams":"best_bigrams.csv",
|
| 120 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 121 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 122 |
+
}
|
| 123 |
+
},
|
| 124 |
+
|
| 125 |
+
|
| 126 |
+
"classes": ['atypical ductal hyperplasia','atypical lobular hyperplasia','atypical papilloma','columnar cell change with atypia','flat epithelial atypia','hyperplasia with atypia','intraductal papilloma','lobular carcinoma in situ','microscopic papilloma','radial scar']
|
| 127 |
+
},
|
| 128 |
+
|
| 129 |
+
"bll_config": {
|
| 130 |
+
"model_option": {
|
| 131 |
+
"single_tfidf": {
|
| 132 |
+
"path_model":"../models/all_labels_hierarchy/single_tfidf/classifiers",
|
| 133 |
+
"model": "bll_xgboost_classifier.pkl",
|
| 134 |
+
"path_vectorizer":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 135 |
+
"vectorizer":"vectorizer_all_branches.pkl",
|
| 136 |
+
"path_bigrmas":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 137 |
+
"bigrams":"best_bigrams.csv",
|
| 138 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 139 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 140 |
+
},
|
| 141 |
+
|
| 142 |
+
"branch_tfidf": {
|
| 143 |
+
"path_model":"../models/all_labels_hierarchy/branch_tfidf/classifiers",
|
| 144 |
+
"model": "bll_xgboost_classifier.pkl",
|
| 145 |
+
"path_vectorizer":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 146 |
+
"vectorizer":"bll_vectorizer.pkl",
|
| 147 |
+
"path_bigrmas":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 148 |
+
"bigrams":"best_bigrams.csv",
|
| 149 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 150 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 151 |
+
}
|
| 152 |
+
},
|
| 153 |
+
|
| 154 |
+
|
| 155 |
+
"classes": ['atypical phyllodes', 'granular cell tumor', 'mucocele']
|
| 156 |
+
},
|
| 157 |
+
|
| 158 |
+
"benign_config": {
|
| 159 |
+
"model_option": {
|
| 160 |
+
"single_tfidf": {
|
| 161 |
+
"path_model":"../models/all_labels_hierarchy/single_tfidf/classifiers",
|
| 162 |
+
"model": "benign_xgboost_classifier.pkl",
|
| 163 |
+
"path_vectorizer":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 164 |
+
"vectorizer":"vectorizer_all_branches.pkl",
|
| 165 |
+
"path_bigrmas":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 166 |
+
"bigrams":"best_bigrams.csv",
|
| 167 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 168 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 169 |
+
},
|
| 170 |
+
|
| 171 |
+
"branch_tfidf": {
|
| 172 |
+
"path_model":"../models/all_labels_hierarchy/branch_tfidf/classifiers",
|
| 173 |
+
"model": "benign_xgboost_classifier.pkl",
|
| 174 |
+
"path_vectorizer":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 175 |
+
"vectorizer":"benign_vectorizer.pkl",
|
| 176 |
+
"path_bigrmas":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 177 |
+
"bigrams":"best_bigrams.csv",
|
| 178 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 179 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 180 |
+
}
|
| 181 |
+
},
|
| 182 |
+
|
| 183 |
+
|
| 184 |
+
"classes": ['apocrine metaplasia','biopsy site changes','columnar cell change without atypia','cyst','excisional or post-surgical change','fat necrosis','fibroadenoma','fibroadenomatoid','fibrocystic disease','fibromatoses','fibrosis','hamartoma','hemangioma','lactational change','lymph node - benign','myofibroblastoma','myxoma','phyllodes','pseudoangiomatous stromal hyperplasia','sclerosing adenosis','usual ductal hyperplasia','fna - benign','seroma']
|
| 185 |
+
},
|
| 186 |
+
|
| 187 |
+
"nbc_config": {
|
| 188 |
+
"model_option": {
|
| 189 |
+
"single_tfidf": {
|
| 190 |
+
"path_model":"../models/all_labels_hierarchy/single_tfidf/classifiers",
|
| 191 |
+
"model": "nbc_xgboost_classifier.pkl",
|
| 192 |
+
"path_vectorizer":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 193 |
+
"vectorizer":"vectorizer_all_branches.pkl",
|
| 194 |
+
"path_bigrmas":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 195 |
+
"bigrams":"best_bigrams.csv",
|
| 196 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/single_tfidf/vectorizers",
|
| 197 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 198 |
+
},
|
| 199 |
+
|
| 200 |
+
"branch_tfidf": {
|
| 201 |
+
"path_model":"../models/all_labels_hierarchy/branch_tfidf/classifiers",
|
| 202 |
+
"model": "nbc_xgboost_classifier.pkl",
|
| 203 |
+
"path_vectorizer":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 204 |
+
"vectorizer":"nbc_vectorizer.pkl",
|
| 205 |
+
"path_bigrmas":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 206 |
+
"bigrams":"best_bigrams.csv",
|
| 207 |
+
"path_phrase_bigrams":"../models/all_labels_hierarchy/branch_tfidf/vectorizers",
|
| 208 |
+
"phrase_bigrams" : "phrase_bigrams.pkl"
|
| 209 |
+
}
|
| 210 |
+
},
|
| 211 |
+
|
| 212 |
+
|
| 213 |
+
"classes": ['lymphoma', 'malignant(sarcomas)', 'non-breast metastasis']
|
| 214 |
+
},
|
| 215 |
+
}
|
| 216 |
+
|
| 217 |
+
return config
|
| 218 |
+
|
| 219 |
+
if __name__ == '__main__':
|
| 220 |
+
pass
|
| 221 |
+
|
app/src/download_models.py
ADDED
|
@@ -0,0 +1,187 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
|
| 2 |
+
""" Download pre-trained models from Google drive. """
|
| 3 |
+
import os
|
| 4 |
+
import argparse
|
| 5 |
+
import zipfile
|
| 6 |
+
import logging
|
| 7 |
+
import requests
|
| 8 |
+
from tqdm import tqdm
|
| 9 |
+
import fire
|
| 10 |
+
import re
|
| 11 |
+
|
| 12 |
+
logging.basicConfig(
|
| 13 |
+
format="%(asctime)s - %(levelname)s - %(filename)s - %(message)s",
|
| 14 |
+
datefmt="%d/%m/%Y %H:%M:%S",
|
| 15 |
+
level=logging.INFO)
|
| 16 |
+
|
| 17 |
+
|
| 18 |
+
"", "", "", "","",""
|
| 19 |
+
|
| 20 |
+
|
| 21 |
+
MODEL_TO_URL = {
|
| 22 |
+
|
| 23 |
+
'PathologyEmoryPubMedBERT': 'https://drive.google.com/open?id=1l_el_mYXoTIQvGwKN2NZbp97E4svH4Fh',
|
| 24 |
+
'PathologyEmoryBERT': 'https://drive.google.com/open?id=11vzo6fJBw1RcdHVBAh6nnn8yua-4kj2IX',
|
| 25 |
+
'ClinicalBERT': 'https://drive.google.com/open?id=1UK9HqSspVneK8zGg7B93vIdTGKK9MI_v',
|
| 26 |
+
'BlueBERT': 'https://drive.google.com/open?id=1o-tcItErOiiwqZ-YRa3sMM3hGB4d3WkP',
|
| 27 |
+
'BioBERT': 'https://drive.google.com/open?id=1m7EkWkFBIBuGbfwg7j0R_WINNnYk3oS9',
|
| 28 |
+
'BERT': 'https://drive.google.com/open?id=1SB_AQAAsHkF79iSAaB3kumYT1rwcOJru',
|
| 29 |
+
|
| 30 |
+
'single_tfidf': 'https://drive.google.com/open?id=1-hxf7sKRtFGMOenlafdkeAr8_9pOz6Ym',
|
| 31 |
+
'branch_tfidf': 'https://drive.google.com/open?id=1pDSnwLFn3YzPRac9rKFV_FN9kdzj2Lb0'
|
| 32 |
+
}
|
| 33 |
+
|
| 34 |
+
"""
|
| 35 |
+
For large Files, Drive requires a Virus Check.
|
| 36 |
+
This function is reponsivle to extract the link from the button confirmation
|
| 37 |
+
"""
|
| 38 |
+
def get_url_from_gdrive_confirmation(contents):
|
| 39 |
+
url = ""
|
| 40 |
+
for line in contents.splitlines():
|
| 41 |
+
m = re.search(r'href="(\/uc\?export=download[^"]+)', line)
|
| 42 |
+
if m:
|
| 43 |
+
url = "https://docs.google.com" + m.groups()[0]
|
| 44 |
+
url = url.replace("&", "&")
|
| 45 |
+
break
|
| 46 |
+
m = re.search('id="downloadForm" action="(.+?)"', line)
|
| 47 |
+
if m:
|
| 48 |
+
url = m.groups()[0]
|
| 49 |
+
url = url.replace("&", "&")
|
| 50 |
+
break
|
| 51 |
+
m = re.search('"downloadUrl":"([^"]+)', line)
|
| 52 |
+
if m:
|
| 53 |
+
url = m.groups()[0]
|
| 54 |
+
url = url.replace("\\u003d", "=")
|
| 55 |
+
url = url.replace("\\u0026", "&")
|
| 56 |
+
break
|
| 57 |
+
m = re.search('<p class="uc-error-subcaption">(.*)</p>', line)
|
| 58 |
+
if m:
|
| 59 |
+
error = m.groups()[0]
|
| 60 |
+
raise RuntimeError(error)
|
| 61 |
+
if not url:
|
| 62 |
+
return None
|
| 63 |
+
return url
|
| 64 |
+
|
| 65 |
+
def download_file_from_google_drive(id, destination):
|
| 66 |
+
URL = "https://docs.google.com/uc?export=download"
|
| 67 |
+
|
| 68 |
+
session = requests.Session()
|
| 69 |
+
|
| 70 |
+
|
| 71 |
+
response = session.get(URL, params={ 'id' : id }, stream=True)
|
| 72 |
+
URL_new = get_url_from_gdrive_confirmation(response.text)
|
| 73 |
+
|
| 74 |
+
if URL_new != None:
|
| 75 |
+
URL = URL_new
|
| 76 |
+
response = session.get(URL, params={ 'id' : id }, stream=True)
|
| 77 |
+
|
| 78 |
+
token = get_confirm_token(response)
|
| 79 |
+
|
| 80 |
+
if token:
|
| 81 |
+
params = { 'id' : id, 'confirm' : token }
|
| 82 |
+
response = session.get(URL, params=params, stream=True)
|
| 83 |
+
|
| 84 |
+
save_response_content(response, destination)
|
| 85 |
+
|
| 86 |
+
def get_confirm_token(response):
|
| 87 |
+
for key, value in response.cookies.items():
|
| 88 |
+
if key.startswith('download_warning'):
|
| 89 |
+
return value
|
| 90 |
+
|
| 91 |
+
return None
|
| 92 |
+
|
| 93 |
+
def save_response_content(response, destination):
|
| 94 |
+
CHUNK_SIZE = 32768
|
| 95 |
+
|
| 96 |
+
with open(destination, "wb") as f:
|
| 97 |
+
for chunk in tqdm(response.iter_content(CHUNK_SIZE)):
|
| 98 |
+
if chunk: # filter out keep-alive new chunks
|
| 99 |
+
f.write(chunk)
|
| 100 |
+
|
| 101 |
+
def check_if_exist(model:str = "single_tfidf"):
|
| 102 |
+
|
| 103 |
+
if model =="single_vectorizer":
|
| 104 |
+
model = "single_tfidf"
|
| 105 |
+
if model =="branch_vectorizer":
|
| 106 |
+
model = "branch_tfidf"
|
| 107 |
+
|
| 108 |
+
project_dir = os.path.dirname(os.path.abspath(__file__))
|
| 109 |
+
if model != None:
|
| 110 |
+
if model in ['single_tfidf', 'branch_tfidf' ]:
|
| 111 |
+
path='models/all_labels_hierarchy/'
|
| 112 |
+
path_model = os.path.join(project_dir, "..", path, model,'classifiers')
|
| 113 |
+
path_vectorizer = os.path.join(project_dir, "..", path, model,'vectorizers')
|
| 114 |
+
if os.path.exists(path_model) and os.path.exists(path_vectorizer):
|
| 115 |
+
if len(os.listdir(path_model)) >0 and len(os.listdir(path_vectorizer)) >0:
|
| 116 |
+
return True
|
| 117 |
+
else:
|
| 118 |
+
path='models/higher_order_hierarchy/'
|
| 119 |
+
path_folder = os.path.join(project_dir, "..", path, model)
|
| 120 |
+
if os.path.exists(path_folder):
|
| 121 |
+
if len(os.listdir(path_folder + "/" )) >1:
|
| 122 |
+
return True
|
| 123 |
+
return False
|
| 124 |
+
|
| 125 |
+
def download_model(all_labels='single_tfidf', higher_order='PathologyEmoryPubMedBERT'):
|
| 126 |
+
project_dir = os.path.dirname(os.path.abspath(__file__))
|
| 127 |
+
|
| 128 |
+
path_all_labels='models/all_labels_hierarchy/'
|
| 129 |
+
path_higher_order='models/higher_order_hierarchy/'
|
| 130 |
+
|
| 131 |
+
def extract_model(path_file, name):
|
| 132 |
+
|
| 133 |
+
os.makedirs(os.path.join(project_dir, "..", path_file), exist_ok=True)
|
| 134 |
+
|
| 135 |
+
file_destination = os.path.join(project_dir, "..", path_file, name + '.zip')
|
| 136 |
+
|
| 137 |
+
file_id = MODEL_TO_URL[name].split('id=')[-1]
|
| 138 |
+
|
| 139 |
+
logging.info(f'Downloading {name} model (~1000MB tar.xz archive)')
|
| 140 |
+
download_file_from_google_drive(file_id, file_destination)
|
| 141 |
+
|
| 142 |
+
logging.info('Extracting model from archive (~1300MB folder) and saving to ' + str(file_destination))
|
| 143 |
+
with zipfile.ZipFile(file_destination, 'r') as zip_ref:
|
| 144 |
+
zip_ref.extractall(path=os.path.dirname(file_destination))
|
| 145 |
+
|
| 146 |
+
logging.info('Removing archive')
|
| 147 |
+
os.remove(file_destination)
|
| 148 |
+
logging.info('Done.')
|
| 149 |
+
|
| 150 |
+
|
| 151 |
+
if higher_order != None:
|
| 152 |
+
if not check_if_exist(higher_order):
|
| 153 |
+
extract_model(path_higher_order, higher_order)
|
| 154 |
+
else:
|
| 155 |
+
logging.info('Model ' + str(higher_order) + ' already exist')
|
| 156 |
+
|
| 157 |
+
if all_labels!= None:
|
| 158 |
+
if not check_if_exist(all_labels):
|
| 159 |
+
extract_model(path_all_labels, all_labels)
|
| 160 |
+
else:
|
| 161 |
+
logging.info('Model ' + str(all_labels) + ' already exist')
|
| 162 |
+
|
| 163 |
+
|
| 164 |
+
|
| 165 |
+
|
| 166 |
+
def download(all_labels:str = "single_tfidf", higher_order:str = "PathologyEmoryPubMedBERT"):
|
| 167 |
+
"""
|
| 168 |
+
Input Options:
|
| 169 |
+
all_labels : single_tfidf, branch_tfidf
|
| 170 |
+
higher_order : clinicalBERT, blueBERT, patho_clinicalBERT, patho_blueBERT, charBERT
|
| 171 |
+
"""
|
| 172 |
+
all_labels_options = [ "single_tfidf", "branch_tfidf"]
|
| 173 |
+
higher_order_option = [ "PathologyEmoryPubMedBERT", "PathologyEmoryBERT", "ClinicalBERT", "BlueBERT","BioBERT","BERT" ]
|
| 174 |
+
|
| 175 |
+
if all_labels not in all_labels_options or higher_order not in higher_order_option:
|
| 176 |
+
print("\n\tPlease provide a valid model for downloading")
|
| 177 |
+
print("\n\t\tall_labels: " + " ".join(x for x in all_labels_options))
|
| 178 |
+
print("\n\t\thigher_order: " + " ".join(x for x in higher_order))
|
| 179 |
+
exit()
|
| 180 |
+
|
| 181 |
+
download_model(all_labels,higher_order)
|
| 182 |
+
|
| 183 |
+
if __name__ == "__main__":
|
| 184 |
+
fire.Fire(download)
|
| 185 |
+
|
| 186 |
+
|
| 187 |
+
|
app/src/label_extraction.py
ADDED
|
@@ -0,0 +1,150 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import pandas as pd
|
| 2 |
+
import numpy as np
|
| 3 |
+
from tqdm import tqdm
|
| 4 |
+
import re
|
| 5 |
+
import fire
|
| 6 |
+
import json
|
| 7 |
+
from tqdm import tqdm
|
| 8 |
+
import logging
|
| 9 |
+
from pipeline import Pipeline
|
| 10 |
+
import copy
|
| 11 |
+
from download_models import check_if_exist
|
| 12 |
+
|
| 13 |
+
"""
|
| 14 |
+
Install dependecies by running: pip3 install -r requirements.txt
|
| 15 |
+
|
| 16 |
+
Running command example:
|
| 17 |
+
python3 label_extraction.py --path_to_file data.xlsx --column_name report --save_predictions predictions.xlsx --save_json output.json
|
| 18 |
+
"""
|
| 19 |
+
|
| 20 |
+
def data_extraction(path_to_file:str, column_name:str, higher_model:str="clinicalBERT", all_label_model="single_tfidf", save_predictions:str=None, output_model_data=None,save_input=None, save_json:str=None):
|
| 21 |
+
|
| 22 |
+
"""
|
| 23 |
+
This program takes an excell/csv sheet and extract the higher order and cancer characteristics from pathology reports
|
| 24 |
+
|
| 25 |
+
Input Options:
|
| 26 |
+
1) path_to_file - Path to an excel/csv with pathology diagnosis: String (Required)
|
| 27 |
+
2) column_name - Which column has the pathology diagnosis: String (Required)
|
| 28 |
+
3) higher_model - Which version of higher order model to use: String (Required)
|
| 29 |
+
4) all_label_model - Which version of all labels model to use: String (Required)
|
| 30 |
+
5) save_predictions - Path to save output: String (Optional)
|
| 31 |
+
6) output_model_data - Option to output model data to csv True/False (Optional)
|
| 32 |
+
7) save_input - Option to output the input fields True/False (Optional)
|
| 33 |
+
8) save_json - Path to save json analyis: String (Optional)
|
| 34 |
+
|
| 35 |
+
|
| 36 |
+
"""
|
| 37 |
+
|
| 38 |
+
data_orig = read_data(path_to_file)
|
| 39 |
+
data_orig = data_orig.fillna("NA")
|
| 40 |
+
data = data_orig.loc[:, ~data_orig.columns.str.contains('^Unnamed')][column_name].values
|
| 41 |
+
|
| 42 |
+
predictions, json_output, higher_order_pred,all_labels_pred = {},[],[],[]
|
| 43 |
+
|
| 44 |
+
if not check_if_exist(higher_model):
|
| 45 |
+
print("\n\t ##### Please Download Model: " + str(higher_model) + "#####")
|
| 46 |
+
exit()
|
| 47 |
+
if not check_if_exist(all_label_model):
|
| 48 |
+
print("\n\t ##### Please Download Model: " + str(all_label_model) + "#####")
|
| 49 |
+
exit()
|
| 50 |
+
|
| 51 |
+
model = Pipeline(bert_option=higher_model, branch_option=all_label_model)
|
| 52 |
+
|
| 53 |
+
logging.info("\nRunning Predictions for data size of: " + str(len(data)))
|
| 54 |
+
for index in tqdm(range(len(data))):
|
| 55 |
+
d = data[index]
|
| 56 |
+
# refactor json
|
| 57 |
+
preds,all_layer_hidden_states = model.run(d)
|
| 58 |
+
predictions["sample_" + str(index)] = {}
|
| 59 |
+
for ind,pred in enumerate(preds):
|
| 60 |
+
predictions["sample_" + str(index)]["prediction_" + str(ind)] = pred
|
| 61 |
+
|
| 62 |
+
for key,sample in predictions.items():
|
| 63 |
+
higher,all_p = [],[]
|
| 64 |
+
for key,pred in sample.items():
|
| 65 |
+
for higher_order, sub_arr in pred.items():
|
| 66 |
+
higher.append(higher_order)
|
| 67 |
+
for label,v in sub_arr['labels'].items():
|
| 68 |
+
all_p.append(label)
|
| 69 |
+
|
| 70 |
+
higher_order_pred.append(" && ".join(x for x in higher))
|
| 71 |
+
all_labels_pred.append(" && ".join(x for x in all_p))
|
| 72 |
+
|
| 73 |
+
|
| 74 |
+
predictions_refact = copy.deepcopy(predictions)
|
| 75 |
+
transformer_data, discriminator_data= [0 for x in range(len(data))], [0 for x in range(len(data))]
|
| 76 |
+
|
| 77 |
+
for index in tqdm(range(len(data))):
|
| 78 |
+
key = "sample_" + str(index)
|
| 79 |
+
for k,v in predictions[key].items():
|
| 80 |
+
for k_s, v_s in v.items():
|
| 81 |
+
predictions_refact["sample_" + str(index)]["data"] = v_s['data']
|
| 82 |
+
predictions_refact["sample_" + str(index)]["transformer_data"] = v_s['transformer_data']
|
| 83 |
+
predictions_refact["sample_" + str(index)]["discriminator_data"] = v_s['word_analysis']['discriminator_data']
|
| 84 |
+
transformer_data[index] = v_s['transformer_data']
|
| 85 |
+
discriminator_data[index] = v_s['word_analysis']['discriminator_data']
|
| 86 |
+
|
| 87 |
+
del predictions_refact[key][k][k_s]['data']
|
| 88 |
+
del predictions_refact[key][k][k_s]['transformer_data']
|
| 89 |
+
del predictions_refact[key][k][k_s]['word_analysis']['discriminator_data']
|
| 90 |
+
|
| 91 |
+
json_output = predictions_refact
|
| 92 |
+
|
| 93 |
+
|
| 94 |
+
if save_predictions!= None:
|
| 95 |
+
logging.info("Saving Predictions")
|
| 96 |
+
if output_model_data != None:
|
| 97 |
+
all_preds = pd.DataFrame(list(zip(higher_order_pred, all_labels_pred,transformer_data,discriminator_data,data)), columns =['Higher Order',"All Labels", 'Higher Order Model Data','All Labels Model Data',column_name])
|
| 98 |
+
else:
|
| 99 |
+
all_preds = pd.DataFrame(list(zip(higher_order_pred, all_labels_pred)), columns =['Higher Order',"All Labels"])
|
| 100 |
+
|
| 101 |
+
if save_input != None:
|
| 102 |
+
all_preds = pd.concat([data_orig, all_preds], axis=1)
|
| 103 |
+
try:
|
| 104 |
+
all_preds.to_excel(save_predictions)
|
| 105 |
+
except ValueError:
|
| 106 |
+
try:
|
| 107 |
+
all_preds.to_csv(save_predictions)
|
| 108 |
+
except ValueError:
|
| 109 |
+
logging.exception("Error while saving predictions " + str(e))
|
| 110 |
+
exit()
|
| 111 |
+
logging.info("Done")
|
| 112 |
+
|
| 113 |
+
if save_json!= None:
|
| 114 |
+
logging.info("Saving Json")
|
| 115 |
+
try:
|
| 116 |
+
with open(save_json, 'w') as f:
|
| 117 |
+
for k, v in json_output.items():
|
| 118 |
+
f.write('{'+str(k) + ':'+ str(v) + '\n')
|
| 119 |
+
|
| 120 |
+
except ValueError:
|
| 121 |
+
logging.exception("Error while saving json analysis " + str(e))
|
| 122 |
+
exit()
|
| 123 |
+
logging.info("Done")
|
| 124 |
+
|
| 125 |
+
|
| 126 |
+
def read_data(path_to_file):
|
| 127 |
+
|
| 128 |
+
try:
|
| 129 |
+
df = pd.read_excel(path_to_file)
|
| 130 |
+
return df
|
| 131 |
+
except ValueError:
|
| 132 |
+
try:
|
| 133 |
+
df = pd.read_csv(path_to_file)
|
| 134 |
+
return df
|
| 135 |
+
except ValueError:
|
| 136 |
+
logging.exception("### Error occurred while splitting document. Info: " + str(e))
|
| 137 |
+
exit()
|
| 138 |
+
|
| 139 |
+
|
| 140 |
+
|
| 141 |
+
def run():
|
| 142 |
+
fire.Fire(data_extraction)
|
| 143 |
+
|
| 144 |
+
if __name__ == '__main__':
|
| 145 |
+
logging.basicConfig(format="%(asctime)s - %(levelname)s - %(filename)s - %(message)s",datefmt="%d/%m/%Y %H:%M:%S",level=logging.INFO)
|
| 146 |
+
run()
|
| 147 |
+
|
| 148 |
+
|
| 149 |
+
|
| 150 |
+
|
app/src/pipeline.py
ADDED
|
@@ -0,0 +1,670 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import sys
|
| 3 |
+
|
| 4 |
+
import text_cleaning_transforerms as tc
|
| 5 |
+
import text_cleaning
|
| 6 |
+
|
| 7 |
+
import logging
|
| 8 |
+
import torch
|
| 9 |
+
|
| 10 |
+
import matplotlib.pyplot as plt
|
| 11 |
+
import numpy as np
|
| 12 |
+
import pandas as pd
|
| 13 |
+
import itertools
|
| 14 |
+
import json
|
| 15 |
+
import joblib
|
| 16 |
+
from gensim.models import phrases
|
| 17 |
+
|
| 18 |
+
import math
|
| 19 |
+
|
| 20 |
+
import xgboost
|
| 21 |
+
import re
|
| 22 |
+
import nltk
|
| 23 |
+
nltk.download('stopwords')
|
| 24 |
+
nltk.download('wordnet')
|
| 25 |
+
import html
|
| 26 |
+
|
| 27 |
+
from config import config_file
|
| 28 |
+
|
| 29 |
+
|
| 30 |
+
from lime import lime_text
|
| 31 |
+
from lime.lime_text import LimeTextExplainer
|
| 32 |
+
|
| 33 |
+
|
| 34 |
+
from transformers import AutoModelForSequenceClassification,AutoTokenizer
|
| 35 |
+
|
| 36 |
+
from nltk.tokenize import word_tokenize
|
| 37 |
+
|
| 38 |
+
|
| 39 |
+
"""
|
| 40 |
+
Cancer Severity Class.
|
| 41 |
+
|
| 42 |
+
export env_name="path"
|
| 43 |
+
"""
|
| 44 |
+
class BERT_Model(object):
|
| 45 |
+
def __init__(self, config,bert_option:str="clinicalBERT"):
|
| 46 |
+
|
| 47 |
+
try:
|
| 48 |
+
self.config = config
|
| 49 |
+
self.project_dir = os.path.dirname(os.path.abspath(__file__))
|
| 50 |
+
self.bert_option = bert_option
|
| 51 |
+
# check if a path was alreadey added to os env table
|
| 52 |
+
|
| 53 |
+
if "model_folder" in os.environ:
|
| 54 |
+
self.config['model_folder'] = os.environ['model_folder']
|
| 55 |
+
else:
|
| 56 |
+
self.config['model_folder'] = os.path.join(self.project_dir, self.config['model_option'][self.bert_option]['model_folder'])
|
| 57 |
+
|
| 58 |
+
self.initialize()
|
| 59 |
+
except Exception as e:
|
| 60 |
+
logging.exception("Error occurred while Initializing BERT Model, please double check you have a config file " +" Info: " + str(e))
|
| 61 |
+
exit()
|
| 62 |
+
|
| 63 |
+
def initialize(self):
|
| 64 |
+
# Set up logging
|
| 65 |
+
logging.basicConfig(
|
| 66 |
+
format="%(asctime)s - %(levelname)s - %(filename)s - %(message)s",
|
| 67 |
+
datefmt="%d/%m/%Y %H:%M:%S",
|
| 68 |
+
level=logging.INFO)
|
| 69 |
+
|
| 70 |
+
# Check for GPUs
|
| 71 |
+
if torch.cuda.is_available():
|
| 72 |
+
self.config["use_cuda"] = True
|
| 73 |
+
self.config["cuda_device"] = torch.cuda.current_device()
|
| 74 |
+
logging.info("Using GPU (`%s`)", torch.cuda.get_device_name())
|
| 75 |
+
else:
|
| 76 |
+
self.config["use_cuda"] = False
|
| 77 |
+
self.config["cuda_device"] = "cpu"
|
| 78 |
+
logging.info("Using CPU")
|
| 79 |
+
|
| 80 |
+
|
| 81 |
+
self.model = AutoModelForSequenceClassification.from_pretrained(self.config["model_folder"], num_labels=len(self.config['classes']),output_hidden_states=True).to(self.config["cuda_device"])
|
| 82 |
+
self.tokenizer = AutoTokenizer.from_pretrained(self.config["model_folder"])
|
| 83 |
+
|
| 84 |
+
|
| 85 |
+
def clean_data(self,text:str):
|
| 86 |
+
return tc.pre_process(text,max_size=int(self.config["max_seq_length"]),remove_punctuation=True )
|
| 87 |
+
|
| 88 |
+
def sigmoid(self,x):
|
| 89 |
+
return 1 / (1 + math.exp(-x))
|
| 90 |
+
|
| 91 |
+
"""
|
| 92 |
+
Convert output of multi-class to probabilities between 0-1
|
| 93 |
+
"""
|
| 94 |
+
def raw_to_probs(self,vector):
|
| 95 |
+
return [self.sigmoid(x) for x in vector]
|
| 96 |
+
|
| 97 |
+
|
| 98 |
+
"""
|
| 99 |
+
Given a threshold, convert a vector of probabiities into predictions (0 or 1)
|
| 100 |
+
"""
|
| 101 |
+
def _threshold(self, vector:list, threshold:float=0.5) -> list:
|
| 102 |
+
logit_vector = [1 if x >=threshold else 0 for x in vector]
|
| 103 |
+
return logit_vector
|
| 104 |
+
|
| 105 |
+
"""
|
| 106 |
+
Pre-Process the data according to the same strategy used during training
|
| 107 |
+
"""
|
| 108 |
+
def pre_process(self,texts:list)-> list:
|
| 109 |
+
transformer_clean_data,transformer_clean_data_chunks = [],[]
|
| 110 |
+
for index,t in enumerate(texts):
|
| 111 |
+
clean_data, clean_data_chunks = self.clean_data(t)
|
| 112 |
+
transformer_clean_data.append(clean_data)
|
| 113 |
+
transformer_clean_data_chunks.append(clean_data_chunks)
|
| 114 |
+
|
| 115 |
+
return transformer_clean_data,transformer_clean_data_chunks
|
| 116 |
+
|
| 117 |
+
|
| 118 |
+
"""
|
| 119 |
+
Giving a list of texts, return the sentence embedding (CLS token from last BERT layer)
|
| 120 |
+
"""
|
| 121 |
+
def get_embeddings(self,texts:list)-> list:
|
| 122 |
+
|
| 123 |
+
transformer_clean_data,_ = self.pre_process(texts)
|
| 124 |
+
|
| 125 |
+
inputs = self.tokenizer(transformer_clean_data, return_tensors="pt", padding=True).to(self.config["cuda_device"])
|
| 126 |
+
outputs = self.model(**inputs,output_hidden_states=True)
|
| 127 |
+
last_hidden_states = outputs[1][-1].detach().cpu().numpy()
|
| 128 |
+
embeddings_output = np.asarray(last_hidden_states[:, 0])
|
| 129 |
+
|
| 130 |
+
return embeddings_output
|
| 131 |
+
|
| 132 |
+
"""
|
| 133 |
+
Giving a list of texts, run BERT prediction for each sample
|
| 134 |
+
If use_chunks is set to True (default), it chunks de data into chunks of max_size (set on config.py)
|
| 135 |
+
The final prediction for that sample is the concatenation of predictions from every chunck
|
| 136 |
+
|
| 137 |
+
Returns:
|
| 138 |
+
* Predictions
|
| 139 |
+
* Probabiities
|
| 140 |
+
* Sentence Embedding (CLS token from last BERT layer)
|
| 141 |
+
* Pre-Processed data used for Prediction
|
| 142 |
+
"""
|
| 143 |
+
def predict(self,texts:list, use_chunks=True)-> list:
|
| 144 |
+
|
| 145 |
+
transformer_clean_data,transformer_clean_data_chunks = self.pre_process(texts)
|
| 146 |
+
print(transformer_clean_data,transformer_clean_data_chunks)
|
| 147 |
+
ids_chunks = []
|
| 148 |
+
# Flat all chunks (2d list) into 1d List (each chunck is feed separetly to prediction)
|
| 149 |
+
if use_chunks:
|
| 150 |
+
|
| 151 |
+
flatten_chunks = [j for sub in transformer_clean_data_chunks for j in sub]
|
| 152 |
+
ids = [[x]*len(transformer_clean_data_chunks[x]) for x in range(len(transformer_clean_data_chunks))]
|
| 153 |
+
ids_chunks = [j for sub in ids for j in sub]
|
| 154 |
+
data = flatten_chunks.copy()
|
| 155 |
+
else:
|
| 156 |
+
data = transformer_clean_data.copy()
|
| 157 |
+
|
| 158 |
+
print(data)
|
| 159 |
+
inputs = self.tokenizer(data, return_tensors="pt", padding=True).to(self.config["cuda_device"])
|
| 160 |
+
outputs = self.model(**inputs,output_hidden_states=True)
|
| 161 |
+
|
| 162 |
+
# Post-Process output if using chunks --> Merge chunck Predictions into 1
|
| 163 |
+
if use_chunks:
|
| 164 |
+
raw_probs_chunks = outputs[0].detach().cpu().numpy()
|
| 165 |
+
probs_chunks = [self.raw_to_probs(x) for x in raw_probs_chunks]
|
| 166 |
+
probs = np.asarray([[0 for x in range(len(probs_chunks[0]))] for x in range(len(texts))],dtype=float)
|
| 167 |
+
for index, prob in enumerate(probs_chunks):
|
| 168 |
+
id_ = ids_chunks[index]
|
| 169 |
+
|
| 170 |
+
# if no predictions for such index yet, add (this is the base - avoid zero preds)
|
| 171 |
+
if np.sum(probs[id_])<=0:
|
| 172 |
+
probs[id_] = prob
|
| 173 |
+
else: # update to merge predictions
|
| 174 |
+
pred = np.asarray(self._threshold(vector=prob,threshold=self.config["threshold_prediction"]))
|
| 175 |
+
pos_pred_index = np.where(pred>0)[0]
|
| 176 |
+
if len(pos_pred_index)>0:
|
| 177 |
+
for pos in pos_pred_index:
|
| 178 |
+
probs[id_][pos] = prob[pos]
|
| 179 |
+
|
| 180 |
+
else:
|
| 181 |
+
raw_probs = outputs[0].detach().cpu().numpy()
|
| 182 |
+
probs = [self.raw_to_probs(x) for x in raw_probs]
|
| 183 |
+
|
| 184 |
+
predictions = [self._threshold(vector=pred,threshold=self.config["threshold_prediction"]) for pred in probs]
|
| 185 |
+
|
| 186 |
+
|
| 187 |
+
|
| 188 |
+
last_hidden_states = outputs[1][-1].detach().cpu().numpy()
|
| 189 |
+
embeddings_output = np.asarray(last_hidden_states[:, 0])
|
| 190 |
+
|
| 191 |
+
return predictions, probs, embeddings_output, transformer_clean_data
|
| 192 |
+
|
| 193 |
+
|
| 194 |
+
|
| 195 |
+
"""
|
| 196 |
+
Giving a list of text, it executes the branch prediction
|
| 197 |
+
This function call BERT Predict, pre-process predictions, and return the post-process branch prediction
|
| 198 |
+
Returns:
|
| 199 |
+
* Branch Prediction
|
| 200 |
+
* Sentence Embedding (CLS token from last BERT layer)
|
| 201 |
+
"""
|
| 202 |
+
def branch_prediction(self,texts:list)-> list:
|
| 203 |
+
out_pred = []
|
| 204 |
+
|
| 205 |
+
predictions, probs, embeddings_output, transformer_clean_data = self.predict(texts,use_chunks=True)
|
| 206 |
+
|
| 207 |
+
try:
|
| 208 |
+
for index, preds in enumerate(probs):
|
| 209 |
+
preds = np.asarray(preds)
|
| 210 |
+
pos = np.where(preds > 0.5)[0]
|
| 211 |
+
pred = []
|
| 212 |
+
if len(pos) >0:
|
| 213 |
+
for ind in pos:
|
| 214 |
+
pred.append({self.config['classes'][ind]: {"probability":preds[ind], "data":texts[index], "transformer_data": transformer_clean_data[index] }})
|
| 215 |
+
else:
|
| 216 |
+
pred.append({"No Prediction": {"probability":0, "data":texts[index], "transformer_data": transformer_clean_data[index]}})
|
| 217 |
+
|
| 218 |
+
out_pred.append(pred)
|
| 219 |
+
except Exception as e:
|
| 220 |
+
logging.exception("Error occurred on BERT model prediction" +" Info: " + str(e))
|
| 221 |
+
exit()
|
| 222 |
+
|
| 223 |
+
return out_pred,embeddings_output
|
| 224 |
+
|
| 225 |
+
|
| 226 |
+
"""
|
| 227 |
+
Cancer Diagnose Prediction Class.
|
| 228 |
+
This class is used to load each individual branch classifier
|
| 229 |
+
"""
|
| 230 |
+
class Branch_Classifier(object):
|
| 231 |
+
def __init__(self, config, branch_option:str="single_tfidf"):
|
| 232 |
+
self.config = config
|
| 233 |
+
self.branch_option = branch_option
|
| 234 |
+
self.project_dir = os.path.dirname(os.path.abspath(__file__))
|
| 235 |
+
|
| 236 |
+
try:
|
| 237 |
+
if "path_model" in os.environ:
|
| 238 |
+
self.config['path_model'] = os.environ['path_model']
|
| 239 |
+
else:
|
| 240 |
+
self.config['path_model'] = os.path.join(self.project_dir, self.config['model_option'][self.branch_option]['path_model'])
|
| 241 |
+
|
| 242 |
+
if "path_vectorizer" in os.environ:
|
| 243 |
+
self.config['path_vectorizer'] = os.environ['path_vectorizer']
|
| 244 |
+
else:
|
| 245 |
+
self.config['path_vectorizer'] = os.path.join(self.project_dir, self.config['model_option'][self.branch_option]['path_vectorizer'])
|
| 246 |
+
|
| 247 |
+
if "path_bigrmas" in os.environ:
|
| 248 |
+
self.config['path_bigrmas'] = os.environ['path_bigrmas']
|
| 249 |
+
else:
|
| 250 |
+
self.config['path_bigrmas'] = os.path.join(self.project_dir, self.config['model_option'][self.branch_option]['path_bigrmas'])
|
| 251 |
+
|
| 252 |
+
if "path_phrase_bigrams" in os.environ:
|
| 253 |
+
self.config['path_phrase_bigrams'] = os.environ['path_phrase_bigrams']
|
| 254 |
+
else:
|
| 255 |
+
self.config['path_phrase_bigrams'] = os.path.join(self.project_dir, self.config['model_option'][self.branch_option]['path_phrase_bigrams'])
|
| 256 |
+
|
| 257 |
+
except Exception as e:
|
| 258 |
+
logging.exception("Error occurred while reading config file. Please read config instructions" +" Info: " + str(e))
|
| 259 |
+
exit()
|
| 260 |
+
|
| 261 |
+
self.initialize()
|
| 262 |
+
|
| 263 |
+
|
| 264 |
+
def initialize(self):
|
| 265 |
+
|
| 266 |
+
try:
|
| 267 |
+
self.model = joblib.load(os.path.join(self.config['path_model'],self.config['model_option'][self.branch_option]['model']))
|
| 268 |
+
self.vectorizer = joblib.load(os.path.join(self.config['path_vectorizer'],self.config['model_option'][self.branch_option]['vectorizer']))
|
| 269 |
+
self.good_bigrams = pd.read_csv(os.path.join(self.config["path_bigrmas"],self.config['model_option'][self.branch_option]['bigrams']))['bigram'].to_list()
|
| 270 |
+
self.phrase_bigrams = phrases.Phrases.load(os.path.join(self.config["path_phrase_bigrams"],self.config['model_option'][self.branch_option]['phrase_bigrams']))
|
| 271 |
+
|
| 272 |
+
except Exception as e:
|
| 273 |
+
logging.exception("Error occurred while initializing models and vectorizer" +" Info: " + str(e))
|
| 274 |
+
exit()
|
| 275 |
+
|
| 276 |
+
"""
|
| 277 |
+
Only add specific Bi-grams (Pre-calculated during Training)
|
| 278 |
+
"""
|
| 279 |
+
def clean_bigram(self,data:list)-> list:
|
| 280 |
+
|
| 281 |
+
data_clean = []
|
| 282 |
+
|
| 283 |
+
for word in data:
|
| 284 |
+
if re.search("_",word) == None:
|
| 285 |
+
data_clean.append(word)
|
| 286 |
+
else: # gotta add the word without _ as well
|
| 287 |
+
if word in self.good_bigrams:
|
| 288 |
+
data_clean.append(word)
|
| 289 |
+
else:
|
| 290 |
+
data_clean.append(word.split("_")[0])
|
| 291 |
+
data_clean.append(word.split("_")[1])
|
| 292 |
+
|
| 293 |
+
return np.asarray(data_clean)
|
| 294 |
+
|
| 295 |
+
"""
|
| 296 |
+
Giving a list of text, pre-process and format the data
|
| 297 |
+
"""
|
| 298 |
+
def format_data(self,data:list)-> list:
|
| 299 |
+
try:
|
| 300 |
+
X = text_cleaning.text_cleaning(data, steam=False, lemma=True,single_input=True)[0]
|
| 301 |
+
|
| 302 |
+
### Add Bigrams and keep only the good ones(pre-selected)
|
| 303 |
+
X_bigrmas = self.phrase_bigrams[X]
|
| 304 |
+
data_clean = self.clean_bigram(X_bigrmas)
|
| 305 |
+
X_bigrams_clean = ' '.join(map(str, data_clean))
|
| 306 |
+
pre_processed = self.vectorizer.transform([X_bigrams_clean]).toarray(),X_bigrams_clean
|
| 307 |
+
|
| 308 |
+
except Exception as e:
|
| 309 |
+
logging.exception("Error occurred while formatting and cleaning data" +" Info: " + str(e))
|
| 310 |
+
exit()
|
| 311 |
+
|
| 312 |
+
return pre_processed
|
| 313 |
+
|
| 314 |
+
|
| 315 |
+
def html_escape(self,text):
|
| 316 |
+
return html.escape(text)
|
| 317 |
+
|
| 318 |
+
def predict(self, texts:list)-> list:
|
| 319 |
+
"""
|
| 320 |
+
Steps:
|
| 321 |
+
1) Run the predictions from higher-order
|
| 322 |
+
2) Based on the prediction, activate which brach(es) to send for final prediction (cancer characteristics)
|
| 323 |
+
3) For final prediction, create a word importance HTML for each input
|
| 324 |
+
"""
|
| 325 |
+
out_pred = {'predictions': {}, 'word_analysis':{},}
|
| 326 |
+
|
| 327 |
+
color = "234, 131, 4" # orange
|
| 328 |
+
try:
|
| 329 |
+
for t in texts:
|
| 330 |
+
text_tfidf,clean_data = self.format_data(t)
|
| 331 |
+
probs = self.model.predict_proba(text_tfidf).toarray()
|
| 332 |
+
predictions = self.model.predict(text_tfidf).toarray()
|
| 333 |
+
for index,preds in enumerate(predictions):
|
| 334 |
+
pos = np.where(preds > 0.5)[0]
|
| 335 |
+
pred = []
|
| 336 |
+
if len(pos) >0:
|
| 337 |
+
for ind in pos:
|
| 338 |
+
highlighted_html_text = []
|
| 339 |
+
weigts = self.model.classifiers_[ind].feature_importances_
|
| 340 |
+
word_weights = {}
|
| 341 |
+
words = clean_data.split()
|
| 342 |
+
min_new = 0
|
| 343 |
+
max_new = 100
|
| 344 |
+
min_old = np.min(weigts)
|
| 345 |
+
max_old = np.max(weigts)
|
| 346 |
+
for w in words:
|
| 347 |
+
found = False
|
| 348 |
+
for word, key in self.vectorizer.vocabulary_.items():
|
| 349 |
+
if w == word:
|
| 350 |
+
found = True
|
| 351 |
+
# rescale weights
|
| 352 |
+
weight = ( (max_new - min_new) / (max_old - min_old) * (weigts[key] - max_old) + max_new)
|
| 353 |
+
if weight <0.5:
|
| 354 |
+
weight = 0
|
| 355 |
+
|
| 356 |
+
|
| 357 |
+
if "_" in w: # add for each word
|
| 358 |
+
w1,w2 = w.split("_")
|
| 359 |
+
word_weights[w1] = weight
|
| 360 |
+
word_weights[w2] = weight
|
| 361 |
+
if w2 =="one":
|
| 362 |
+
word_weights["1"] = weight
|
| 363 |
+
word_weights["i"] = weight
|
| 364 |
+
if w2 =="two":
|
| 365 |
+
word_weights["2"] = weight
|
| 366 |
+
word_weights["ii"] = weight
|
| 367 |
+
if w2 =="three":
|
| 368 |
+
word_weights["3"] = weight
|
| 369 |
+
word_weights["iii"] = weight
|
| 370 |
+
else:
|
| 371 |
+
word_weights[w] = weight
|
| 372 |
+
if found == False: # some words aren't presented in the model
|
| 373 |
+
word_weights[w] = 0
|
| 374 |
+
|
| 375 |
+
words = word_tokenize(t.lower().replace("-", " - ").replace("_", " ").replace(".", " . ").replace(",", " , ").replace("(", " ( ").replace(")", " ) "))
|
| 376 |
+
for i,w in enumerate(words):
|
| 377 |
+
if w not in word_weights or w=='-' or w==',' or w=='.' or w=="(" or w==")":
|
| 378 |
+
word_weights[w] = 0
|
| 379 |
+
highlighted_html_text.append(w)
|
| 380 |
+
else:
|
| 381 |
+
weight = 0 if word_weights[w] <1 else word_weights[w]
|
| 382 |
+
highlighted_html_text.append('<span font-size:40px; ; style="background-color:rgba(' + color + ',' + str(weight) + ');">' + self.html_escape(w) + '</span>')
|
| 383 |
+
|
| 384 |
+
|
| 385 |
+
|
| 386 |
+
highlighted_html_text = ' '.join(highlighted_html_text)
|
| 387 |
+
#pred.append({ "predictions": {self.config['classes'][ind]: {"probability":probs[index][ind]}},"word_analysis": {"discriminator_data": clean_data,"word_importance": word_weights, "highlighted_html_text":highlighted_html_text}})
|
| 388 |
+
out_pred["predictions"][self.config['classes'][ind]] = {"probability":probs[index][ind]}
|
| 389 |
+
out_pred["word_analysis"] = {"discriminator_data": clean_data,"word_importance": word_weights, "highlighted_html_text":highlighted_html_text}
|
| 390 |
+
|
| 391 |
+
else:
|
| 392 |
+
out_pred["predictions"] = {"Unkown": {"probability":0.5}}
|
| 393 |
+
out_pred["word_analysis"] = {"discriminator_data": clean_data,"word_importance": {x:0 for x in t.split()}, "highlighted_html_text": " ".join(x for x in t.split())}
|
| 394 |
+
|
| 395 |
+
#pred.append({"predictions": {"Unkown": {"probability":0.5}}, "word_analysis": {"discriminator_data": clean_data,"word_importance": {x:0 for x in t.split()}, "highlighted_html_text": " ".join(x for x in t.split())}})
|
| 396 |
+
|
| 397 |
+
#out_pred.append(pred)
|
| 398 |
+
|
| 399 |
+
except Exception as e:
|
| 400 |
+
logging.exception("Error occurred on model prediction" +" Info: " + str(e))
|
| 401 |
+
exit()
|
| 402 |
+
|
| 403 |
+
return out_pred
|
| 404 |
+
|
| 405 |
+
|
| 406 |
+
class LIME_Interpretability(object):
|
| 407 |
+
|
| 408 |
+
"""
|
| 409 |
+
Class for LIME Analysis
|
| 410 |
+
|
| 411 |
+
"""
|
| 412 |
+
|
| 413 |
+
def __init__(self, label_colors = { "positive": "234, 131, 4", # orange
|
| 414 |
+
"negative":'65, 137, 225', # blue
|
| 415 |
+
}):
|
| 416 |
+
|
| 417 |
+
self.color_classes = label_colors
|
| 418 |
+
|
| 419 |
+
# function to normalize, if applicable
|
| 420 |
+
def __normalize_MinMax(self,arr, t_min=0, t_max=1):
|
| 421 |
+
norm_arr = []
|
| 422 |
+
diff = t_max - t_min
|
| 423 |
+
diff_arr = max(arr) - min(arr)
|
| 424 |
+
for i in arr:
|
| 425 |
+
temp = (((i - min(arr)) * diff) / diff_arr) + t_min
|
| 426 |
+
norm_arr.append(temp)
|
| 427 |
+
return norm_arr
|
| 428 |
+
|
| 429 |
+
|
| 430 |
+
def __html_escape(self,text):
|
| 431 |
+
return html.escape(text)
|
| 432 |
+
|
| 433 |
+
|
| 434 |
+
def __add_bigrams(self,txt):
|
| 435 |
+
fixed_bigrams = [ [' gradeone ', 'grade 1', 'grade i', 'grade I', 'grade one',],
|
| 436 |
+
[' gradetwo ', 'grade 2', 'grade ii', 'grade II', 'grade two', ],
|
| 437 |
+
[' gradethree ', 'grade 3' , 'grade iii', 'grade III', 'grade three']]
|
| 438 |
+
for b in fixed_bigrams:
|
| 439 |
+
sub = ""
|
| 440 |
+
not_first = False
|
| 441 |
+
for x in b[1:]:
|
| 442 |
+
if not_first:
|
| 443 |
+
sub += "|"
|
| 444 |
+
not_first = True
|
| 445 |
+
|
| 446 |
+
sub += str(x) + "|" + str(x) + " " + "|" + " " + str(x) + "|" + " " + str(x)
|
| 447 |
+
txt = re.sub(sub, b[0], txt)
|
| 448 |
+
# Removing multiple spaces
|
| 449 |
+
txt = re.sub(r'\s+', ' ', txt)
|
| 450 |
+
txt = re.sub(' +', ' ', txt)
|
| 451 |
+
return txt
|
| 452 |
+
|
| 453 |
+
def __highlight_full_data(self,lime_weights, data, exp_labels,class_names):
|
| 454 |
+
words_p = [x[0] for x in lime_weights if x[1]>0]
|
| 455 |
+
weights_p = np.asarray([x[1] for x in lime_weights if x[1] >0])
|
| 456 |
+
if len(weights_p) >1:
|
| 457 |
+
weights_p = self.__normalize_MinMax(weights_p, t_min=min(weights_p), t_max=1)
|
| 458 |
+
else:
|
| 459 |
+
weights_p = [1]
|
| 460 |
+
words_n = [x[0] for x in lime_weights if x[1]<0]
|
| 461 |
+
weights_n = np.asarray([x[1] for x in lime_weights if x[1] <0])
|
| 462 |
+
# weights_n = self.__normalize_MinMax(weights_n, t_min=max(weights_p), t_max=-0.8)
|
| 463 |
+
|
| 464 |
+
labels = exp_labels
|
| 465 |
+
pred = class_names[labels[0]]
|
| 466 |
+
corr_pred = class_names[labels[1]] # negative lime weights
|
| 467 |
+
|
| 468 |
+
# positive values
|
| 469 |
+
df_coeff = pd.DataFrame(
|
| 470 |
+
{'word': words_p,
|
| 471 |
+
'num_code': weights_p
|
| 472 |
+
})
|
| 473 |
+
word_to_coeff_mapping_p = {}
|
| 474 |
+
for row in df_coeff.iterrows():
|
| 475 |
+
row = row[1]
|
| 476 |
+
word_to_coeff_mapping_p[row[0]] = row[1]
|
| 477 |
+
|
| 478 |
+
# negative values
|
| 479 |
+
df_coeff = pd.DataFrame(
|
| 480 |
+
{'word': words_n,
|
| 481 |
+
'num_code': weights_n
|
| 482 |
+
})
|
| 483 |
+
|
| 484 |
+
word_to_coeff_mapping_n = {}
|
| 485 |
+
for row in df_coeff.iterrows():
|
| 486 |
+
row = row[1]
|
| 487 |
+
word_to_coeff_mapping_n[row[0]] = row[1]
|
| 488 |
+
|
| 489 |
+
max_alpha = 1
|
| 490 |
+
highlighted_text = []
|
| 491 |
+
data = re.sub("-"," ", data)
|
| 492 |
+
data = re.sub("/","", data)
|
| 493 |
+
for word in word_tokenize(self.__add_bigrams(data)):
|
| 494 |
+
if word.lower() in word_to_coeff_mapping_p or word.lower() in word_to_coeff_mapping_n:
|
| 495 |
+
if word.lower() in word_to_coeff_mapping_p:
|
| 496 |
+
weight = word_to_coeff_mapping_p[word.lower()]
|
| 497 |
+
else:
|
| 498 |
+
weight = word_to_coeff_mapping_n[word.lower()]
|
| 499 |
+
|
| 500 |
+
if weight >0:
|
| 501 |
+
color = self.color_classes["positive"]
|
| 502 |
+
else:
|
| 503 |
+
color = self.color_classes["negative"]
|
| 504 |
+
weight *= -1
|
| 505 |
+
weight *=10
|
| 506 |
+
|
| 507 |
+
highlighted_text.append('<span font-size:40px; ; style="background-color:rgba(' + color + ',' + str(weight) + ');">' + self.__html_escape(word) + '</span>')
|
| 508 |
+
|
| 509 |
+
else:
|
| 510 |
+
highlighted_text.append(word)
|
| 511 |
+
|
| 512 |
+
highlighted_text = ' '.join(highlighted_text)
|
| 513 |
+
|
| 514 |
+
return highlighted_text
|
| 515 |
+
|
| 516 |
+
|
| 517 |
+
def lime_analysis(self,model,data_original, data_clean, num_features=30, num_samples=50, top_labels=2,
|
| 518 |
+
class_names=['ibc', 'nbc', 'isc', 'bll', 'hrl', 'benign', 'negative']):
|
| 519 |
+
|
| 520 |
+
# LIME Predictor Function
|
| 521 |
+
def predict(texts):
|
| 522 |
+
results = []
|
| 523 |
+
for text in texts:
|
| 524 |
+
predictions, probs, embeddings_output, transformer_clean_data = model.predict([text],use_chunks=False)
|
| 525 |
+
results.append(probs[0])
|
| 526 |
+
|
| 527 |
+
return np.array(results)
|
| 528 |
+
|
| 529 |
+
explainer = LimeTextExplainer(class_names=class_names)
|
| 530 |
+
exp = explainer.explain_instance(data_clean, predict, num_features=num_features,
|
| 531 |
+
num_samples=num_samples, top_labels=top_labels)
|
| 532 |
+
l = exp.available_labels()
|
| 533 |
+
run_info = exp.as_list(l[0])
|
| 534 |
+
return self.__highlight_full_data(run_info, data_original, l,class_names)
|
| 535 |
+
|
| 536 |
+
|
| 537 |
+
"""
|
| 538 |
+
The pipeline is responsible to consolidate the output of all models (higher order and all labels hierarchy)
|
| 539 |
+
It takes a string as input, and returns a jason with higher-order(Severity) and all labels(Diagnose) predictions and their probability score
|
| 540 |
+
"""
|
| 541 |
+
class Pipeline(object):
|
| 542 |
+
|
| 543 |
+
def __init__(self, bert_option:str="clinicalBERT", branch_option:str="single_tfidf"):
|
| 544 |
+
logging.basicConfig(format="%(asctime)s - %(levelname)s - %(filename)s - %(message)s",datefmt="%d/%m/%Y %H:%M:%S",level=logging.INFO)
|
| 545 |
+
|
| 546 |
+
if branch_option =="single_vectorizer":
|
| 547 |
+
self.branch_option = "single_tfidf"
|
| 548 |
+
elif branch_option =="branch_vectorizer":
|
| 549 |
+
self.branch_option = "branch_tfidf"
|
| 550 |
+
else:
|
| 551 |
+
self.branch_option=branch_option
|
| 552 |
+
|
| 553 |
+
self.bert_option=bert_option
|
| 554 |
+
|
| 555 |
+
try:
|
| 556 |
+
self.config = config_file()
|
| 557 |
+
self.BERT_config = self.config['BERT_config']
|
| 558 |
+
self.ibc_config = self.config['ibc_config']
|
| 559 |
+
self.isc_config = self.config['isc_config']
|
| 560 |
+
self.hrl_config = self.config['hrl_config']
|
| 561 |
+
self.bll_config = self.config['bll_config']
|
| 562 |
+
self.benign_config = self.config['benign_config']
|
| 563 |
+
self.nbc_config = self.config['nbc_config']
|
| 564 |
+
|
| 565 |
+
except Exception as e:
|
| 566 |
+
logging.exception("Error occurred while initializing models and vectorizer" +" Info: " + str(e))
|
| 567 |
+
exit()
|
| 568 |
+
|
| 569 |
+
self.lime_interpretability = LIME_Interpretability()
|
| 570 |
+
|
| 571 |
+
self.initialize()
|
| 572 |
+
|
| 573 |
+
|
| 574 |
+
def initialize(self):
|
| 575 |
+
try:
|
| 576 |
+
self.bert_model = BERT_Model(self.BERT_config, self.bert_option)
|
| 577 |
+
try:
|
| 578 |
+
self.ibc_branch = Branch_Classifier(self.ibc_config,branch_option=self.branch_option)
|
| 579 |
+
except Exception as e:
|
| 580 |
+
logging.exception("Error occurred while Initializing IBC branch Model, please double check you have a config file " +" Info: " + str(e))
|
| 581 |
+
exit()
|
| 582 |
+
|
| 583 |
+
try:
|
| 584 |
+
self.isc_branch = Branch_Classifier(self.isc_config,branch_option=self.branch_option)
|
| 585 |
+
except Exception as e:
|
| 586 |
+
logging.exception("Error occurred while Initializing isc branch Model, please double check you have a config file " +" Info: " + str(e))
|
| 587 |
+
exit()
|
| 588 |
+
|
| 589 |
+
try:
|
| 590 |
+
self.hrl_branch = Branch_Classifier(self.hrl_config,branch_option=self.branch_option)
|
| 591 |
+
except Exception as e:
|
| 592 |
+
logging.exception("Error occurred while Initializing hrl branch Model, please double check you have a config file " +" Info: " + str(e))
|
| 593 |
+
exit()
|
| 594 |
+
|
| 595 |
+
try:
|
| 596 |
+
self.bll_branch = Branch_Classifier(self.bll_config,branch_option=self.branch_option)
|
| 597 |
+
except Exception as e:
|
| 598 |
+
logging.exception("Error occurred while Initializing bll branch Model, please double check you have a config file " +" Info: " + str(e))
|
| 599 |
+
exit()
|
| 600 |
+
|
| 601 |
+
try:
|
| 602 |
+
self.benign_branch = Branch_Classifier(self.benign_config,branch_option=self.branch_option)
|
| 603 |
+
except Exception as e:
|
| 604 |
+
logging.exception("Error occurred while Initializing benign branch Model, please double check you have a config file " +" Info: " + str(e))
|
| 605 |
+
exit()
|
| 606 |
+
|
| 607 |
+
try:
|
| 608 |
+
self.nbc_branch = Branch_Classifier(self.nbc_config,branch_option=self.branch_option)
|
| 609 |
+
except Exception as e:
|
| 610 |
+
logging.exception("Error occurred while Initializing nbc branch Model, please double check you have a config file " +" Info: " + str(e))
|
| 611 |
+
exit()
|
| 612 |
+
|
| 613 |
+
self.all_label_models = [self.ibc_branch,self.nbc_branch,self.isc_branch,self.bll_branch,self.hrl_branch,self.benign_branch]
|
| 614 |
+
|
| 615 |
+
|
| 616 |
+
except Exception as e:
|
| 617 |
+
logging.exception("Error occurred while Initializing Pipeline, please double check you have a config file " +" Info: " + str(e))
|
| 618 |
+
exit()
|
| 619 |
+
|
| 620 |
+
|
| 621 |
+
"""
|
| 622 |
+
Run the entire pipeline
|
| 623 |
+
Steps:
|
| 624 |
+
1) First, we run the Severity Prediction (BERT)
|
| 625 |
+
2) Given each prediction for each sample, we then:
|
| 626 |
+
2.1) Run the corresponding Diagnose Branch Prediction
|
| 627 |
+
2.2) Merge every branch prediction
|
| 628 |
+
3) Merge Every Severity and Branch Prediction
|
| 629 |
+
|
| 630 |
+
Inputs:
|
| 631 |
+
* Text
|
| 632 |
+
|
| 633 |
+
Output:
|
| 634 |
+
* Predictions (Predictions + Probabilites)
|
| 635 |
+
* Sentence Embedding
|
| 636 |
+
"""
|
| 637 |
+
def run(self,input_text:str):
|
| 638 |
+
|
| 639 |
+
"""
|
| 640 |
+
First, get the severity prediction (higher order branch)
|
| 641 |
+
"""
|
| 642 |
+
predictions,embeddings_output = self.bert_model.branch_prediction([input_text])
|
| 643 |
+
predictions = predictions[0]
|
| 644 |
+
for pred in predictions:
|
| 645 |
+
for higher_order, sub_arr in pred.items():
|
| 646 |
+
# Check which branch it belongs to
|
| 647 |
+
if higher_order in ["Negative","No Prediction"]:
|
| 648 |
+
pred[higher_order]['labels'] = {higher_order: {"probability":sub_arr['probability']}}
|
| 649 |
+
pred[higher_order]["word_analysis"] = {"discriminator_data": "Not Used", "word_importance": {x:0 for x in input_text.split()}, "highlighted_html_text": " ".join(x for x in input_text.split())}
|
| 650 |
+
|
| 651 |
+
# For each Severity, run the corresponding Branch Prediction
|
| 652 |
+
else:
|
| 653 |
+
model = self.all_label_models[self.bert_model.config['classes'].index(higher_order)]
|
| 654 |
+
out_pred = model.predict([input_text])
|
| 655 |
+
|
| 656 |
+
pred[higher_order]['labels'] = out_pred['predictions']
|
| 657 |
+
pred[higher_order]['word_analysis'] = out_pred['word_analysis']
|
| 658 |
+
|
| 659 |
+
return predictions,embeddings_output
|
| 660 |
+
|
| 661 |
+
def bert_interpretability(self, input_text:str):
|
| 662 |
+
return self.lime_interpretability.lime_analysis(self.bert_model,input_text, self.bert_model.clean_data(input_text), class_names=self.bert_model.config['classes'])
|
| 663 |
+
|
| 664 |
+
|
| 665 |
+
if __name__ == '__main__':
|
| 666 |
+
exit()
|
| 667 |
+
|
| 668 |
+
|
| 669 |
+
|
| 670 |
+
|
app/src/text_cleaning.py
ADDED
|
@@ -0,0 +1,250 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from gensim.parsing import preprocessing
|
| 2 |
+
from gensim.parsing.preprocessing import strip_tags, strip_punctuation,strip_numeric,remove_stopwords
|
| 3 |
+
import re
|
| 4 |
+
from nltk.stem import PorterStemmer
|
| 5 |
+
import nltk
|
| 6 |
+
from nltk.corpus import stopwords
|
| 7 |
+
import pandas as pd
|
| 8 |
+
|
| 9 |
+
def remove_noise_text(txt):
|
| 10 |
+
|
| 11 |
+
txt = txt.lower()
|
| 12 |
+
txt = re.sub('right|left', '', txt) # remove right/left spaces
|
| 13 |
+
txt = re.sub("primary site:", '', txt)
|
| 14 |
+
|
| 15 |
+
#txt = re.sub('post-surgical changes', ' ', txt.lower())
|
| 16 |
+
|
| 17 |
+
# Remove any mentions to " Findings were discussed with...."
|
| 18 |
+
txt = txt.split("findings were discussed with")[0]
|
| 19 |
+
|
| 20 |
+
# Remove any other occurance of PI's Information
|
| 21 |
+
txt = txt.split("this study has been reviewed and interpreted")[0]
|
| 22 |
+
txt = txt.split("this finding was communicated to")[0]
|
| 23 |
+
txt = txt.split("important findings were identified")[0]
|
| 24 |
+
txt = txt.split("these findings")[0]
|
| 25 |
+
txt = txt.split("findings above were")[0]
|
| 26 |
+
txt = txt.split("findings regarding")[0]
|
| 27 |
+
txt = txt.split("were discussed")[0]
|
| 28 |
+
txt = txt.split("these images were")[0]
|
| 29 |
+
txt = txt.split("important finding")[0]
|
| 30 |
+
|
| 31 |
+
# remove any section headers
|
| 32 |
+
txt = re.sub("post-surgical changes:", '', txt)
|
| 33 |
+
txt = re.sub("post surgical changes:", '', txt)
|
| 34 |
+
txt = re.sub("primary site:", '', txt)
|
| 35 |
+
txt = re.sub("primary site", '', txt)
|
| 36 |
+
txt = re.sub("neck:", '', txt)
|
| 37 |
+
txt = re.sub("post-treatment changes:", '', txt)
|
| 38 |
+
txt = re.sub("post treatment changes:", '', txt)
|
| 39 |
+
txt = re.sub("brain, orbits, spine and lungs:", '', txt)
|
| 40 |
+
txt = re.sub("primary :", '', txt)
|
| 41 |
+
txt = re.sub("neck:", '', txt)
|
| 42 |
+
txt = re.sub("aerodigestive tract:", '', txt)
|
| 43 |
+
txt = re.sub("calvarium, skull base, and spine:", '', txt)
|
| 44 |
+
txt = re.sub("other:", '', txt)
|
| 45 |
+
txt = re.sub("upper neck:", '', txt)
|
| 46 |
+
txt = re.sub("perineural disease:", '', txt)
|
| 47 |
+
txt = re.sub("technique:", '', txt)
|
| 48 |
+
txt = re.sub("comparison:", '', txt)
|
| 49 |
+
txt = re.sub("paranasal sinuses:", '', txt)
|
| 50 |
+
txt = re.sub("included orbits:", '', txt)
|
| 51 |
+
txt = re.sub("nasopharynx:", '', txt)
|
| 52 |
+
txt = re.sub("tympanomastoid cavities:", '', txt)
|
| 53 |
+
txt = re.sub("skull base and calvarium:", '', txt)
|
| 54 |
+
txt = re.sub("included intracranial structures:", '', txt)
|
| 55 |
+
txt = re.sub("abnormal enhancement:", '', txt)
|
| 56 |
+
txt = re.sub("lymph nodes:", '', txt)
|
| 57 |
+
txt = re.sub("impression:", '', txt)
|
| 58 |
+
txt = re.sub("nodes:", '', txt)
|
| 59 |
+
txt = re.sub("mri orbits:", '', txt)
|
| 60 |
+
txt = re.sub("mri brain:", '', txt)
|
| 61 |
+
txt = re.sub("brain:", '', txt)
|
| 62 |
+
txt = re.sub("ct face w/:", '', txt)
|
| 63 |
+
txt = re.sub("transspatial extension:", '', txt)
|
| 64 |
+
txt = re.sub("thyroid bed:", '', txt)
|
| 65 |
+
txt = re.sub("additional findings:", '', txt)
|
| 66 |
+
txt = re.sub("series_image", '', txt)
|
| 67 |
+
txt = re.sub("series image", '', txt)
|
| 68 |
+
txt = re.sub("image series", '', txt)
|
| 69 |
+
txt = re.sub("series", '', txt)
|
| 70 |
+
|
| 71 |
+
txt = re.sub(" mm | mm|mm ", " ", txt)
|
| 72 |
+
txt = re.sub(" series | series|series ", "", txt)
|
| 73 |
+
txt = re.sub(" cm | cm|cm ", " ", txt)
|
| 74 |
+
txt = re.sub(" cc | cc|cc ", " ", txt)
|
| 75 |
+
txt = re.sub(" ct | ct|ct ", " ", txt)
|
| 76 |
+
txt = re.sub(" mri | mri|mri ", " ", txt)
|
| 77 |
+
txt = re.sub(" see | see|see ", " ", txt)
|
| 78 |
+
txt = re.sub(" iia | iia|iia ", " ", txt)
|
| 79 |
+
txt = re.sub("comment", "", txt)
|
| 80 |
+
|
| 81 |
+
|
| 82 |
+
txt = re.sub("post treatment", '', txt)
|
| 83 |
+
txt = re.sub("post_treatment", '', txt)
|
| 84 |
+
txt = re.sub("post-treatment", '', txt)
|
| 85 |
+
txt = re.sub("findings suggest", '', txt)
|
| 86 |
+
txt = re.sub("findings", '', txt)
|
| 87 |
+
txt = re.sub("suggest", '', txt)
|
| 88 |
+
txt = re.sub("study reviewed", '', txt)
|
| 89 |
+
txt = re.sub("study", '', txt)
|
| 90 |
+
txt = re.sub("reviewed", '', txt)
|
| 91 |
+
txt = re.sub("please see", '', txt)
|
| 92 |
+
txt = re.sub("please", '', txt)
|
| 93 |
+
|
| 94 |
+
txt = re.sub("skull base", '', txt)
|
| 95 |
+
txt = re.sub("fdg avid", '', txt)
|
| 96 |
+
txt = re.sub("fdg aivity", '', txt)
|
| 97 |
+
txt = re.sub("please see chest ct for further evaluation of known lung mass", '', txt)
|
| 98 |
+
|
| 99 |
+
txt = re.sub("status_post", '', txt)
|
| 100 |
+
txt = re.sub("status post|clock|/|'/'", '', txt)
|
| 101 |
+
txt = re.sub("statuspost|:", '', txt)
|
| 102 |
+
txt = re.sub(" cm | cm|cm ", " centimeters ", txt)
|
| 103 |
+
txt = re.sub(" cc | cc|cc ", " cubic centimeters ", txt)
|
| 104 |
+
txt = re.sub(" ct | ct|ct ", " carat metric ", txt)
|
| 105 |
+
txt = re.sub(" mm | mm|mm ", " millimeters ", txt)
|
| 106 |
+
#txt = re.sub("(\\d*\\.\\d+)|(\\d+\\.[0-9 ]+)","",txt)
|
| 107 |
+
|
| 108 |
+
# in the worst case, just replace the name from PI to empty string
|
| 109 |
+
txt = re.sub("dr\\.\\s[^\\s]+", '', txt)
|
| 110 |
+
|
| 111 |
+
|
| 112 |
+
txt = re.sub('\\;', ' .', txt)
|
| 113 |
+
txt = re.sub('\\.', ' .', txt)
|
| 114 |
+
|
| 115 |
+
# Removing multiple spaces
|
| 116 |
+
txt = re.sub(r'\s+', ' ', txt)
|
| 117 |
+
|
| 118 |
+
|
| 119 |
+
return txt
|
| 120 |
+
|
| 121 |
+
def add_bigrams(txt, fixed_bigrams):
|
| 122 |
+
|
| 123 |
+
for b in fixed_bigrams:
|
| 124 |
+
sub = ""
|
| 125 |
+
not_first = False
|
| 126 |
+
for x in b[1:]:
|
| 127 |
+
if not_first:
|
| 128 |
+
sub += "|"
|
| 129 |
+
not_first = True
|
| 130 |
+
|
| 131 |
+
sub += str(x) + "|" + str(x) + " " + "|" + " " + str(x) + "|" + " " + str(x)
|
| 132 |
+
txt = re.sub(sub, b[0], txt)
|
| 133 |
+
|
| 134 |
+
return txt
|
| 135 |
+
|
| 136 |
+
|
| 137 |
+
def clean_text(txt_orig,filters,stop_words,non_stop_words,freq_words,fixed_bigrams,steam, lemma , clean, min_lenght, eightify=False):
|
| 138 |
+
txt = remove_noise_text(txt_orig)
|
| 139 |
+
|
| 140 |
+
#print("\n\t\tOriginal\n", txt)
|
| 141 |
+
txt = add_bigrams(txt, fixed_bigrams)
|
| 142 |
+
#print("\n\t\tCleaned\n", txt)
|
| 143 |
+
words = preprocessing.preprocess_string(txt, filters)
|
| 144 |
+
words = add_bigrams(" ".join(w for w in words), fixed_bigrams).split()
|
| 145 |
+
|
| 146 |
+
txt = " ".join(w for w in words)
|
| 147 |
+
|
| 148 |
+
# eightify
|
| 149 |
+
#
|
| 150 |
+
if eightify:
|
| 151 |
+
replaces = [ ["her2|her 2|her two", " hertwo "], ["0", "8"], ["1", "8"], ["2", "8"], ["3", "8"],["4", "8"],
|
| 152 |
+
["5", "8"],["6", "8"] ,["7", "8"] ,["8", "8"] ,["9", "8"] ,
|
| 153 |
+
["\\>", " greather "], ["\\<", " less "]]
|
| 154 |
+
|
| 155 |
+
else:
|
| 156 |
+
replaces = [ ["her2|her 2|her two", " hertwo "], ["0", "zero "], ["1", "one "], ["2", "two "], ["3", "three "],["4", "four "],
|
| 157 |
+
["5", "five "],["6", "six "] ,["7", "seven "] ,["8", "eight "] ,["9", "nine " ] ,
|
| 158 |
+
["\\>", " greather "], ["\\<", " less "]]
|
| 159 |
+
|
| 160 |
+
|
| 161 |
+
for sub in replaces:
|
| 162 |
+
txt = re.sub(sub[0], sub[1], txt)
|
| 163 |
+
|
| 164 |
+
# Removing multiple spaces
|
| 165 |
+
txt = re.sub(r'\s+', ' ', txt)
|
| 166 |
+
|
| 167 |
+
words = txt.split()
|
| 168 |
+
|
| 169 |
+
if clean:
|
| 170 |
+
words = [w for w in words if (not w in stop_words and re.search("[a-z-A-Z]+\\w+",w) != None and (len(w) >min_lenght or w in non_stop_words) or w=='.') ]
|
| 171 |
+
else:
|
| 172 |
+
words = [w for w in words if (re.search("[a-z-A-Z]+\\w+",w) != None and (len(w) >min_lenght or w in non_stop_words) or w=='.')]
|
| 173 |
+
|
| 174 |
+
c_words = words.copy()
|
| 175 |
+
|
| 176 |
+
if steam:
|
| 177 |
+
porter = PorterStemmer()
|
| 178 |
+
c_words = [porter.stem(word) for word in c_words if not porter.stem(word) in freq_words and (len(porter.stem(word)) >min_lenght or word in non_stop_words or word=='.')]
|
| 179 |
+
|
| 180 |
+
if lemma:
|
| 181 |
+
lem = nltk.stem.wordnet.WordNetLemmatizer()
|
| 182 |
+
c_words = [lem.lemmatize(word) for word in c_words if not lem.lemmatize(word) in freq_words and (len(lem.lemmatize(word)) >min_lenght or word in non_stop_words or word=='.')]
|
| 183 |
+
|
| 184 |
+
return c_words
|
| 185 |
+
|
| 186 |
+
|
| 187 |
+
def text_cleaning(data, steam=False, lemma = True, clean=True, min_lenght=2, remove_punctuation=True,
|
| 188 |
+
freq_words_analysis=False, single_input=False,eightify=True):
|
| 189 |
+
|
| 190 |
+
clean_txt = []
|
| 191 |
+
|
| 192 |
+
|
| 193 |
+
freq_words = ["breast","biopsy","margin","dual","tissue","excision","change","core","identified",
|
| 194 |
+
"mastectomy","site","report","lesion","superior","anterior","inferior","medial",
|
| 195 |
+
"lateral","synoptic","evidence","slide", "brbx"]
|
| 196 |
+
|
| 197 |
+
# position 0 means the bigram output - 1:end means how they may come on text
|
| 198 |
+
fixed_bigrams = [ [' grade_one ', 'grade 1', 'grade i', 'grade I', 'grade one',],
|
| 199 |
+
[' grade_two ', 'grade 2', 'grade ii', 'grade II', 'grade two', ],
|
| 200 |
+
[' grade_three ', 'grade 3' , 'grade iii', 'grade III', 'grade three']]
|
| 201 |
+
|
| 202 |
+
|
| 203 |
+
if remove_punctuation:
|
| 204 |
+
filters = [lambda x: x.lower(), strip_tags, strip_punctuation]
|
| 205 |
+
else:
|
| 206 |
+
filters = [lambda x: x.lower(), strip_tags]
|
| 207 |
+
|
| 208 |
+
stop_words = set(stopwords.words('english'))
|
| 209 |
+
non_stop_words = ['no', 'than', 'not']
|
| 210 |
+
for x in non_stop_words:
|
| 211 |
+
stop_words.remove(x)
|
| 212 |
+
|
| 213 |
+
if single_input:
|
| 214 |
+
c_words = clean_text(data,filters,stop_words,non_stop_words,freq_words,fixed_bigrams,steam, lemma, clean, min_lenght,eightify=eightify)
|
| 215 |
+
if len(c_words)>0:
|
| 216 |
+
if c_words[0] =='.':
|
| 217 |
+
c_words = c_words[1:]
|
| 218 |
+
clean_txt.append(c_words)
|
| 219 |
+
|
| 220 |
+
else:
|
| 221 |
+
for i in range(data.shape[0]):
|
| 222 |
+
txt_orig = data.iloc[i].lower()
|
| 223 |
+
c_words = clean_text(txt_orig,filters,stop_words,non_stop_words,freq_words,fixed_bigrams,steam, lemma, clean, min_lenght,eightify=eightify)
|
| 224 |
+
if len(c_words)>0:
|
| 225 |
+
if c_words[0] =='.':
|
| 226 |
+
c_words = c_words[1:]
|
| 227 |
+
clean_txt.append(c_words)
|
| 228 |
+
|
| 229 |
+
|
| 230 |
+
if freq_words_analysis:
|
| 231 |
+
flatten_corpus = [j for sub in clean_txt for j in sub]
|
| 232 |
+
clean_txt = []
|
| 233 |
+
unique = list(set(flatten_corpus))
|
| 234 |
+
wordfreq = [flatten_corpus.count(p) for p in unique]
|
| 235 |
+
wordfreq = dict(list(zip(unique,wordfreq)))
|
| 236 |
+
|
| 237 |
+
freqdict = [(wordfreq[key], key) for key in wordfreq]
|
| 238 |
+
freqdict.sort()
|
| 239 |
+
freqdict.reverse()
|
| 240 |
+
|
| 241 |
+
df = pd.DataFrame(freqdict,columns = ['Frequency','Word'])
|
| 242 |
+
|
| 243 |
+
|
| 244 |
+
df.to_excel('../mammo_word_count.xls')
|
| 245 |
+
|
| 246 |
+
return clean_txt
|
| 247 |
+
|
| 248 |
+
if __name__ == '__main__':
|
| 249 |
+
exit()
|
| 250 |
+
|
app/src/text_cleaning_transforerms.py
ADDED
|
@@ -0,0 +1,229 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import pandas as pd
|
| 2 |
+
from os import walk
|
| 3 |
+
from os import listdir
|
| 4 |
+
from os.path import isfile, join
|
| 5 |
+
import numpy as np
|
| 6 |
+
import re
|
| 7 |
+
|
| 8 |
+
from gensim.parsing import preprocessing
|
| 9 |
+
from gensim.parsing.preprocessing import strip_tags, strip_punctuation
|
| 10 |
+
from nltk.tokenize import word_tokenize, sent_tokenize
|
| 11 |
+
import math
|
| 12 |
+
from tqdm import tqdm
|
| 13 |
+
|
| 14 |
+
def remove_noise_text(txt):
|
| 15 |
+
|
| 16 |
+
txt = txt.lower()
|
| 17 |
+
txt = re.sub("primary site:", ' ', txt)
|
| 18 |
+
|
| 19 |
+
#txt = re.sub('post-surgical changes', ' ', txt.lower())
|
| 20 |
+
|
| 21 |
+
# Remove any mentions to " Findings were discussed with...."
|
| 22 |
+
txt = txt.split("findings were discussed with")[0]
|
| 23 |
+
|
| 24 |
+
# Remove any other occurance of PI's Information
|
| 25 |
+
txt = txt.split("this study has been reviewed and interpreted")[0]
|
| 26 |
+
txt = txt.split("this finding was communicated to")[0]
|
| 27 |
+
txt = txt.split("important findings were identified")[0]
|
| 28 |
+
txt = txt.split("these findings")[0]
|
| 29 |
+
txt = txt.split("findings above were")[0]
|
| 30 |
+
txt = txt.split("findings regarding")[0]
|
| 31 |
+
txt = txt.split("were discussed")[0]
|
| 32 |
+
txt = txt.split("these images were")[0]
|
| 33 |
+
txt = txt.split("important finding")[0]
|
| 34 |
+
|
| 35 |
+
# remove any section headers
|
| 36 |
+
txt = re.sub("post-surgical changes:", ' ', txt)
|
| 37 |
+
txt = re.sub("post surgical changes:", ' ', txt)
|
| 38 |
+
txt = re.sub("primary site:", ' ', txt)
|
| 39 |
+
txt = re.sub("primary site", ' ', txt)
|
| 40 |
+
txt = re.sub("neck:", ' ', txt)
|
| 41 |
+
txt = re.sub("post-treatment changes:", ' ', txt)
|
| 42 |
+
txt = re.sub("post treatment changes:", ' ', txt)
|
| 43 |
+
txt = re.sub("brain, orbits, spine and lungs:", ' ', txt)
|
| 44 |
+
txt = re.sub("primary :", ' ', txt)
|
| 45 |
+
txt = re.sub("neck:", ' ', txt)
|
| 46 |
+
txt = re.sub("aerodigestive tract:", ' ', txt)
|
| 47 |
+
txt = re.sub("calvarium, skull base, and spine:", ' ', txt)
|
| 48 |
+
txt = re.sub("other:", ' ', txt)
|
| 49 |
+
txt = re.sub("upper neck:", ' ', txt)
|
| 50 |
+
txt = re.sub("perineural disease:", ' ', txt)
|
| 51 |
+
txt = re.sub("technique:", ' ', txt)
|
| 52 |
+
txt = re.sub("comparison:", ' ', txt)
|
| 53 |
+
txt = re.sub("paranasal sinuses:", ' ', txt)
|
| 54 |
+
txt = re.sub("included orbits:", ' ', txt)
|
| 55 |
+
txt = re.sub("nasopharynx:", ' ', txt)
|
| 56 |
+
txt = re.sub("tympanomastoid cavities:", ' ', txt)
|
| 57 |
+
txt = re.sub("skull base and calvarium:", ' ', txt)
|
| 58 |
+
txt = re.sub("included intracranial structures:", ' ', txt)
|
| 59 |
+
txt = re.sub("impression:", ' ', txt)
|
| 60 |
+
txt = re.sub("nodes:", ' ', txt)
|
| 61 |
+
txt = re.sub("mri orbits:", ' ', txt)
|
| 62 |
+
txt = re.sub("mri brain:", ' ', txt)
|
| 63 |
+
txt = re.sub("brain:", ' ', txt)
|
| 64 |
+
txt = re.sub("ct face w/:", ' ', txt)
|
| 65 |
+
txt = re.sub("transspatial extension:", ' ', txt)
|
| 66 |
+
txt = re.sub("thyroid bed:", ' ', txt)
|
| 67 |
+
txt = re.sub("additional findings:", ' ', txt)
|
| 68 |
+
txt = re.sub("series_image", ' ', txt)
|
| 69 |
+
txt = re.sub("series image", ' ', txt)
|
| 70 |
+
txt = re.sub("image series", ' ', txt)
|
| 71 |
+
txt = re.sub("see synoptic report", ' ', txt)
|
| 72 |
+
txt = re.sub("see report", ' ', txt)
|
| 73 |
+
|
| 74 |
+
txt = re.sub("brstwo|brstmarun|brstwln|brlump|lnbx", ' ', txt)
|
| 75 |
+
|
| 76 |
+
txt = re.sub("post_treatment", 'post treatment', txt)
|
| 77 |
+
txt = re.sub("post-treatment", 'post treatment', txt)
|
| 78 |
+
|
| 79 |
+
txt = re.sub("nonmasslike", 'non mass like', txt)
|
| 80 |
+
txt = re.sub("non_mass_like", 'non mass like', txt)
|
| 81 |
+
txt = re.sub("non-mass-like", 'non mass like', txt)
|
| 82 |
+
txt = re.sub("statuspost", 'status post', txt)
|
| 83 |
+
|
| 84 |
+
|
| 85 |
+
# in the worst case, just replace the name from PI to empty string
|
| 86 |
+
txt = re.sub("dr\\.\\s[^\\s]+", ' ', txt)
|
| 87 |
+
|
| 88 |
+
txt = re.sub(" series | series|series ", "", txt)
|
| 89 |
+
txt = re.sub(" cm | cm|cm ", " centimeters ", txt)
|
| 90 |
+
txt = re.sub(" cc | cc|cc ", " cubic centimeters ", txt)
|
| 91 |
+
txt = re.sub(" ct | ct|ct ", " carat metric ", txt)
|
| 92 |
+
txt = re.sub(" mm | mm|mm ", " millimeters ", txt)
|
| 93 |
+
|
| 94 |
+
txt = re.sub("status_post|o\'", '', txt)
|
| 95 |
+
txt = re.sub("status post|clock|/|'/'", '', txt)
|
| 96 |
+
txt = re.sub("statuspost", '', txt)
|
| 97 |
+
txt = re.sub("brstwo|brlump|brstmarun|brwire|brstcap|", '', txt)
|
| 98 |
+
|
| 99 |
+
txt = re.sub("\\(|\\)", ',', txt)
|
| 100 |
+
txt = re.sub(",,", ',', txt)
|
| 101 |
+
txt = re.sub(",\\.", '.', txt)
|
| 102 |
+
txt = re.sub(", \\.", '.', txt)
|
| 103 |
+
|
| 104 |
+
txt = re.sub(" ,", ', ', txt)
|
| 105 |
+
txt = re.sub("a\\.", ' ', txt[0:5]) + txt[5:]
|
| 106 |
+
txt = re.sub("b\\.", ' ', txt[0:5]) + txt[5:]
|
| 107 |
+
txt = re.sub("c\\.", ' ', txt[0:5]) + txt[5:]
|
| 108 |
+
txt = re.sub("d\\.", ' ', txt[0:5]) + txt[5:]
|
| 109 |
+
txt = re.sub("e\\.", ' ', txt[0:5]) + txt[5:]
|
| 110 |
+
txt = re.sub("f\\.", ' ', txt[0:5]) + txt[5:]
|
| 111 |
+
|
| 112 |
+
|
| 113 |
+
# in the worst case, just replace the name from PI to empty string
|
| 114 |
+
txt = re.sub("dr\\.\\s[^\\s]+", '', txt)
|
| 115 |
+
|
| 116 |
+
# Removing multiple spaces
|
| 117 |
+
txt = re.sub(r'\s+', ' ', txt)
|
| 118 |
+
txt = re.sub(' +', ' ', txt)
|
| 119 |
+
|
| 120 |
+
txt = txt.rstrip().lstrip()
|
| 121 |
+
|
| 122 |
+
return txt
|
| 123 |
+
|
| 124 |
+
|
| 125 |
+
def add_bigrams(txt, fixed_bigrams):
|
| 126 |
+
|
| 127 |
+
for b in fixed_bigrams:
|
| 128 |
+
sub = ""
|
| 129 |
+
not_first = False
|
| 130 |
+
for x in b[1:]:
|
| 131 |
+
if not_first:
|
| 132 |
+
sub += "|"
|
| 133 |
+
not_first = True
|
| 134 |
+
|
| 135 |
+
sub += str(x) + "|" + str(x) + " " + "|" + " " + str(x) + "|" + " " + str(x)
|
| 136 |
+
txt = re.sub(sub, b[0], txt)
|
| 137 |
+
|
| 138 |
+
|
| 139 |
+
return txt
|
| 140 |
+
|
| 141 |
+
def extra_clean_text(clean_t,fixed_bigrams):
|
| 142 |
+
|
| 143 |
+
txt = add_bigrams(clean_t, fixed_bigrams)
|
| 144 |
+
replaces = [ ["her2|her 2|her two", " hertwo "],
|
| 145 |
+
# ["0", "zero "], ["1", "one "], ["2", "two "], ["3", "three "],["4", "four "],
|
| 146 |
+
# ["5", "five "],["6", "six "] ,["7", "seven "] ,["8", "eight "] ,["9", "nine " ] ,
|
| 147 |
+
["\\>", " greather "], ["\\<", " less "]]
|
| 148 |
+
|
| 149 |
+
for sub in replaces:
|
| 150 |
+
txt = re.sub(sub[0], sub[1], txt)
|
| 151 |
+
|
| 152 |
+
return txt
|
| 153 |
+
|
| 154 |
+
|
| 155 |
+
def text_cleaning(data,min_lenght=2,extra_clean=True, remove_punctuation=False):
|
| 156 |
+
|
| 157 |
+
# position 0 means the bigram output - 1:end means how they may come on text
|
| 158 |
+
fixed_bigrams = [ [' gradeone ', 'grade 1', 'grade i', 'grade I', 'grade one',],
|
| 159 |
+
[' gradetwo ', 'grade 2', 'grade ii', 'grade II', 'grade two', ],
|
| 160 |
+
[' gradethree ', 'grade 3' , 'grade iii', 'grade III', 'grade three']]
|
| 161 |
+
|
| 162 |
+
clean_txt = []
|
| 163 |
+
|
| 164 |
+
clean_t = remove_noise_text(data)
|
| 165 |
+
if extra_clean:
|
| 166 |
+
clean_t = extra_clean_text(clean_t,fixed_bigrams)
|
| 167 |
+
if remove_punctuation:
|
| 168 |
+
filters = [lambda x: x.lower(), strip_tags, strip_punctuation]
|
| 169 |
+
else:
|
| 170 |
+
filters = [lambda x: x.lower(), strip_tags]
|
| 171 |
+
|
| 172 |
+
clean_t = " ".join(x for x in preprocessing.preprocess_string(clean_t, filters) if len(x) >=min_lenght)
|
| 173 |
+
|
| 174 |
+
|
| 175 |
+
# Removing multiple spaces
|
| 176 |
+
clean_t = re.sub(r'\s+', ' ', clean_t)
|
| 177 |
+
|
| 178 |
+
return clean_t
|
| 179 |
+
|
| 180 |
+
# set only_data = True if no need to get scores or if dataaset doesn't have a score
|
| 181 |
+
def pre_process(data,min_lenght=2,max_size=64, extra_clean=True, remove_punctuation=False):
|
| 182 |
+
|
| 183 |
+
|
| 184 |
+
data_pre_processed = text_cleaning(data,min_lenght=min_lenght,extra_clean=extra_clean, remove_punctuation=remove_punctuation)
|
| 185 |
+
|
| 186 |
+
"""
|
| 187 |
+
Partion the data into max_size chunks
|
| 188 |
+
"""
|
| 189 |
+
sentences = sent_tokenize(data)
|
| 190 |
+
data_pre_processed_chunks,sample = [],""
|
| 191 |
+
|
| 192 |
+
# Were able to split into sentences
|
| 193 |
+
if len(sentences)>1:
|
| 194 |
+
for index,sentence in enumerate(sentences):
|
| 195 |
+
if len(sentence.split()) + len(sample.split()) <= max_size:
|
| 196 |
+
sample += sentence
|
| 197 |
+
else:
|
| 198 |
+
data_pre_processed_chunks.append(text_cleaning(sample,min_lenght=min_lenght,extra_clean=extra_clean, remove_punctuation=remove_punctuation))
|
| 199 |
+
sample = sentence if index < len(sentences)-1 else ""
|
| 200 |
+
|
| 201 |
+
if len(sample) ==0:
|
| 202 |
+
clean_data = text_cleaning(sentences[-1],min_lenght=min_lenght,extra_clean=extra_clean, remove_punctuation=remove_punctuation)
|
| 203 |
+
else:
|
| 204 |
+
clean_data = text_cleaning(sample,min_lenght=min_lenght,extra_clean=extra_clean, remove_punctuation=remove_punctuation)
|
| 205 |
+
|
| 206 |
+
#if len(clean_data.split()) >3:
|
| 207 |
+
data_pre_processed_chunks.append(clean_data)
|
| 208 |
+
|
| 209 |
+
# Split by get max size chunks
|
| 210 |
+
else:
|
| 211 |
+
words = word_tokenize(data)
|
| 212 |
+
lower_b, upper_b = 0, max_size
|
| 213 |
+
for x in range(math.ceil(len(words)/max_size)):
|
| 214 |
+
sample = " ".join(x for x in words[lower_b:upper_b])
|
| 215 |
+
lower_b, upper_b = upper_b, upper_b+max_size
|
| 216 |
+
clean_data = text_cleaning(sample,min_lenght=min_lenght,extra_clean=extra_clean, remove_punctuation=remove_punctuation)
|
| 217 |
+
#if len(clean_data.split()) >3:
|
| 218 |
+
data_pre_processed_chunks.append(clean_data)
|
| 219 |
+
|
| 220 |
+
# return the pre_processed of whoole text and chunks
|
| 221 |
+
return data_pre_processed,data_pre_processed_chunks
|
| 222 |
+
|
| 223 |
+
if __name__ == '__main__':
|
| 224 |
+
exit(1)
|
| 225 |
+
|
| 226 |
+
|
| 227 |
+
|
| 228 |
+
|
| 229 |
+
|
environment.yml
ADDED
|
@@ -0,0 +1,210 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
name: pathology
|
| 2 |
+
channels:
|
| 3 |
+
- pytorch
|
| 4 |
+
- defaults
|
| 5 |
+
dependencies:
|
| 6 |
+
- _libgcc_mutex=0.1=main
|
| 7 |
+
- _openmp_mutex=4.5=1_gnu
|
| 8 |
+
- blas=1.0=mkl
|
| 9 |
+
- ca-certificates=2021.10.26=h06a4308_2
|
| 10 |
+
- certifi=2021.10.8=py38h06a4308_2
|
| 11 |
+
- cudatoolkit=11.3.1=h2bc3f7f_2
|
| 12 |
+
- intel-openmp=2022.0.1=h06a4308_3633
|
| 13 |
+
- ld_impl_linux-64=2.35.1=h7274673_9
|
| 14 |
+
- libffi=3.3=he6710b0_2
|
| 15 |
+
- libgcc-ng=9.3.0=h5101ec6_17
|
| 16 |
+
- libgomp=9.3.0=h5101ec6_17
|
| 17 |
+
- libstdcxx-ng=9.3.0=hd4cf53a_17
|
| 18 |
+
- libuv=1.40.0=h7b6447c_0
|
| 19 |
+
- mkl=2022.0.1=h06a4308_117
|
| 20 |
+
- ncurses=6.3=h7f8727e_2
|
| 21 |
+
- openssl=1.1.1m=h7f8727e_0
|
| 22 |
+
- pip=21.2.4=py38h06a4308_0
|
| 23 |
+
- python=3.8.12=h12debd9_0
|
| 24 |
+
- pytorch-mutex=1.0=cuda
|
| 25 |
+
- readline=8.1.2=h7f8727e_1
|
| 26 |
+
- setuptools=58.0.4=py38h06a4308_0
|
| 27 |
+
- sqlite=3.37.0=hc218d9a_0
|
| 28 |
+
- tk=8.6.11=h1ccaba5_0
|
| 29 |
+
- typing_extensions=3.10.0.2=pyh06a4308_0
|
| 30 |
+
- wheel=0.37.1=pyhd3eb1b0_0
|
| 31 |
+
- xz=5.2.5=h7b6447c_0
|
| 32 |
+
- zlib=1.2.11=h7f8727e_4
|
| 33 |
+
- pip:
|
| 34 |
+
- absl-py==1.0.0
|
| 35 |
+
- aiohttp==3.8.1
|
| 36 |
+
- aiosignal==1.2.0
|
| 37 |
+
- altair==4.2.0
|
| 38 |
+
- argon2-cffi==21.3.0
|
| 39 |
+
- argon2-cffi-bindings==21.2.0
|
| 40 |
+
- astor==0.8.1
|
| 41 |
+
- asttokens==2.0.5
|
| 42 |
+
- async-timeout==4.0.2
|
| 43 |
+
- attrs==21.4.0
|
| 44 |
+
- backcall==0.2.0
|
| 45 |
+
- backports-zoneinfo==0.2.1
|
| 46 |
+
- base58==2.1.1
|
| 47 |
+
- black==22.1.0
|
| 48 |
+
- bleach==4.1.0
|
| 49 |
+
- blinker==1.4
|
| 50 |
+
- cachetools==5.0.0
|
| 51 |
+
- cffi==1.15.0
|
| 52 |
+
- charset-normalizer==2.0.11
|
| 53 |
+
- click==8.0.3
|
| 54 |
+
- configparser==5.2.0
|
| 55 |
+
- cycler==0.11.0
|
| 56 |
+
- datasets==1.18.2
|
| 57 |
+
- debugpy==1.5.1
|
| 58 |
+
- decorator==5.1.1
|
| 59 |
+
- defusedxml==0.7.1
|
| 60 |
+
- dill==0.3.4
|
| 61 |
+
- docker-pycreds==0.4.0
|
| 62 |
+
- entrypoints==0.3
|
| 63 |
+
- et-xmlfile==1.1.0
|
| 64 |
+
- executing==0.8.2
|
| 65 |
+
- filelock==3.4.2
|
| 66 |
+
- fire==0.4.0
|
| 67 |
+
- fonttools==4.29.0
|
| 68 |
+
- frozenlist==1.3.0
|
| 69 |
+
- fsspec==2022.1.0
|
| 70 |
+
- gensim==4.1.2
|
| 71 |
+
- gitdb==4.0.9
|
| 72 |
+
- gitpython==3.1.26
|
| 73 |
+
- google-auth==2.6.0
|
| 74 |
+
- google-auth-oauthlib==0.4.6
|
| 75 |
+
- grpcio==1.43.0
|
| 76 |
+
- htbuilder==0.6.0
|
| 77 |
+
- huggingface-hub==0.4.0
|
| 78 |
+
- idna==3.3
|
| 79 |
+
- imageio==2.14.1
|
| 80 |
+
- importlib-metadata==4.10.1
|
| 81 |
+
- importlib-resources==5.4.0
|
| 82 |
+
- ipykernel==6.7.0
|
| 83 |
+
- ipython==8.0.1
|
| 84 |
+
- ipython-genutils==0.2.0
|
| 85 |
+
- ipywidgets==7.6.5
|
| 86 |
+
- iteration-utilities==0.11.0
|
| 87 |
+
- jedi==0.18.1
|
| 88 |
+
- jinja2==3.0.3
|
| 89 |
+
- joblib==1.1.0
|
| 90 |
+
- jsonschema==4.4.0
|
| 91 |
+
- jupyter-client==7.1.2
|
| 92 |
+
- jupyter-core==4.9.1
|
| 93 |
+
- jupyterlab-pygments==0.1.2
|
| 94 |
+
- jupyterlab-widgets==1.0.2
|
| 95 |
+
- kiwisolver==1.3.2
|
| 96 |
+
- lime==0.2.0.1
|
| 97 |
+
- llvmlite==0.38.0
|
| 98 |
+
- markdown==3.3.6
|
| 99 |
+
- markupsafe==2.0.1
|
| 100 |
+
- matplotlib==3.5.1
|
| 101 |
+
- matplotlib-inline==0.1.3
|
| 102 |
+
- mistune==0.8.4
|
| 103 |
+
- multidict==6.0.2
|
| 104 |
+
- multiprocess==0.70.12.2
|
| 105 |
+
- mypy-extensions==0.4.3
|
| 106 |
+
- nbclient==0.5.10
|
| 107 |
+
- nbconvert==6.4.1
|
| 108 |
+
- nbformat==5.1.3
|
| 109 |
+
- nest-asyncio==1.5.4
|
| 110 |
+
- networkx==2.6.3
|
| 111 |
+
- nltk==3.6.7
|
| 112 |
+
- notebook==6.4.8
|
| 113 |
+
- numba==0.55.1
|
| 114 |
+
- numpy==1.21.5
|
| 115 |
+
- oauthlib==3.2.0
|
| 116 |
+
- openpyxl==3.0.9
|
| 117 |
+
- packaging==21.3
|
| 118 |
+
- pandas==1.4.0
|
| 119 |
+
- pandocfilters==1.5.0
|
| 120 |
+
- parso==0.8.3
|
| 121 |
+
- pathspec==0.9.0
|
| 122 |
+
- pathtools==0.1.2
|
| 123 |
+
- pexpect==4.8.0
|
| 124 |
+
- pickleshare==0.7.5
|
| 125 |
+
- pillow==9.0.0
|
| 126 |
+
- platformdirs==2.4.1
|
| 127 |
+
- plotly==5.5.0
|
| 128 |
+
- prometheus-client==0.13.1
|
| 129 |
+
- promise==2.3
|
| 130 |
+
- prompt-toolkit==3.0.26
|
| 131 |
+
- protobuf==3.19.4
|
| 132 |
+
- psutil==5.9.0
|
| 133 |
+
- ptyprocess==0.7.0
|
| 134 |
+
- pure-eval==0.2.2
|
| 135 |
+
- pyarrow==6.0.1
|
| 136 |
+
- pyasn1==0.4.8
|
| 137 |
+
- pyasn1-modules==0.2.8
|
| 138 |
+
- pycparser==2.21
|
| 139 |
+
- pydeck==0.7.1
|
| 140 |
+
- pygments==2.11.2
|
| 141 |
+
- pympler==1.0.1
|
| 142 |
+
- pynndescent==0.5.6
|
| 143 |
+
- pyparsing==3.0.7
|
| 144 |
+
- pyrsistent==0.18.1
|
| 145 |
+
- python-dateutil==2.8.2
|
| 146 |
+
- pytz==2021.3
|
| 147 |
+
- pytz-deprecation-shim==0.1.0.post0
|
| 148 |
+
- pywavelets==1.2.0
|
| 149 |
+
- pyyaml==6.0
|
| 150 |
+
- pyzmq==22.3.0
|
| 151 |
+
- regex==2022.1.18
|
| 152 |
+
- requests==2.27.1
|
| 153 |
+
- requests-oauthlib==1.3.1
|
| 154 |
+
- rsa==4.8
|
| 155 |
+
- sacremoses==0.0.47
|
| 156 |
+
- scikit-image==0.19.1
|
| 157 |
+
- scikit-learn==1.0.2
|
| 158 |
+
- scikit-multilearn==0.2.0
|
| 159 |
+
- scipy==1.7.3
|
| 160 |
+
- semantic-version==2.8.5
|
| 161 |
+
- send2trash==1.8.0
|
| 162 |
+
- sentencepiece==0.1.96
|
| 163 |
+
- sentry-sdk==1.5.4
|
| 164 |
+
- seqeval==1.2.2
|
| 165 |
+
- setuptools-rust==1.1.2
|
| 166 |
+
- shortuuid==1.0.8
|
| 167 |
+
- simpletransformers==0.63.4
|
| 168 |
+
- six==1.16.0
|
| 169 |
+
- smart-open==5.2.1
|
| 170 |
+
- smmap==5.0.0
|
| 171 |
+
- stack-data==0.1.4
|
| 172 |
+
- streamlit==1.5.0
|
| 173 |
+
- subprocess32==3.5.4
|
| 174 |
+
- tenacity==8.0.1
|
| 175 |
+
- tensorboard==2.8.0
|
| 176 |
+
- tensorboard-data-server==0.6.1
|
| 177 |
+
- tensorboard-plugin-wit==1.8.1
|
| 178 |
+
- termcolor==1.1.0
|
| 179 |
+
- terminado==0.13.1
|
| 180 |
+
- testpath==0.5.0
|
| 181 |
+
- threadpoolctl==3.1.0
|
| 182 |
+
- tifffile==2021.11.2
|
| 183 |
+
- tokenizers==0.11.4
|
| 184 |
+
- toml==0.10.2
|
| 185 |
+
- tomli==2.0.0
|
| 186 |
+
- toolz==0.11.2
|
| 187 |
+
- torch==1.9.0
|
| 188 |
+
- torchvision==0.10.0
|
| 189 |
+
- tornado==6.1
|
| 190 |
+
- tqdm==4.62.3
|
| 191 |
+
- traitlets==5.1.1
|
| 192 |
+
- transformers==4.16.2
|
| 193 |
+
- tzdata==2021.5
|
| 194 |
+
- tzlocal==4.1
|
| 195 |
+
- umap-learn==0.5.2
|
| 196 |
+
- urllib3==1.26.8
|
| 197 |
+
- validators==0.18.2
|
| 198 |
+
- wandb==0.12.9
|
| 199 |
+
- watchdog==2.1.6
|
| 200 |
+
- wcwidth==0.2.5
|
| 201 |
+
- webencodings==0.5.1
|
| 202 |
+
- werkzeug==2.0.2
|
| 203 |
+
- widgetsnbextension==3.5.2
|
| 204 |
+
- xgboost==1.4.2
|
| 205 |
+
- xxhash==2.0.2
|
| 206 |
+
- yarl==1.7.2
|
| 207 |
+
- yaspin==2.1.0
|
| 208 |
+
- zipp==3.7.0
|
| 209 |
+
|
| 210 |
+
|
requirements.txt
ADDED
|
@@ -0,0 +1,180 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Flask==1.1.2
|
| 2 |
+
Flask_Login==0.5.0
|
| 3 |
+
Flask_SQLAlchemy==2.5.1
|
| 4 |
+
Flask_WTF==0.15.1
|
| 5 |
+
WTForms==2.3.3
|
| 6 |
+
absl-py==1.0.0
|
| 7 |
+
aiohttp==3.8.1
|
| 8 |
+
aiosignal==1.2.0
|
| 9 |
+
altair==4.2.0
|
| 10 |
+
argon2-cffi==21.3.0
|
| 11 |
+
argon2-cffi-bindings==21.2.0
|
| 12 |
+
astor==0.8.1
|
| 13 |
+
asttokens==2.0.5
|
| 14 |
+
async-timeout==4.0.2
|
| 15 |
+
attrs==21.4.0
|
| 16 |
+
backcall==0.2.0
|
| 17 |
+
backports-zoneinfo==0.2.1
|
| 18 |
+
base58==2.1.1
|
| 19 |
+
black==22.1.0
|
| 20 |
+
bleach==4.1.0
|
| 21 |
+
blinker==1.4
|
| 22 |
+
cachetools==5.0.0
|
| 23 |
+
cffi==1.15.0
|
| 24 |
+
charset-normalizer==2.0.11
|
| 25 |
+
click==8.0.3
|
| 26 |
+
configparser==5.2.0
|
| 27 |
+
cycler==0.11.0
|
| 28 |
+
datasets==1.18.2
|
| 29 |
+
debugpy==1.5.1
|
| 30 |
+
decorator==5.1.1
|
| 31 |
+
defusedxml==0.7.1
|
| 32 |
+
dill==0.3.4
|
| 33 |
+
docker-pycreds==0.4.0
|
| 34 |
+
entrypoints==0.3
|
| 35 |
+
et-xmlfile==1.1.0
|
| 36 |
+
executing==0.8.2
|
| 37 |
+
filelock==3.4.2
|
| 38 |
+
fire==0.4.0
|
| 39 |
+
fonttools==4.29.0
|
| 40 |
+
frozenlist==1.3.0
|
| 41 |
+
fsspec==2022.1.0
|
| 42 |
+
gensim==4.1.2
|
| 43 |
+
gitdb==4.0.9
|
| 44 |
+
gitpython==3.1.26
|
| 45 |
+
google-auth==2.6.0
|
| 46 |
+
google-auth-oauthlib==0.4.6
|
| 47 |
+
grpcio==1.43.0
|
| 48 |
+
htbuilder==0.6.0
|
| 49 |
+
huggingface-hub==0.4.0
|
| 50 |
+
idna==3.3
|
| 51 |
+
imageio==2.14.1
|
| 52 |
+
importlib-metadata==4.10.1
|
| 53 |
+
importlib-resources==5.4.0
|
| 54 |
+
ipykernel==6.7.0
|
| 55 |
+
ipython==8.0.1
|
| 56 |
+
ipython-genutils==0.2.0
|
| 57 |
+
ipywidgets==7.6.5
|
| 58 |
+
iteration-utilities==0.11.0
|
| 59 |
+
jedi==0.18.1
|
| 60 |
+
jinja2==3.0.3
|
| 61 |
+
joblib==1.1.0
|
| 62 |
+
jsonschema==4.4.0
|
| 63 |
+
jupyter-client==7.1.2
|
| 64 |
+
jupyter-core==4.9.1
|
| 65 |
+
jupyterlab-pygments==0.1.2
|
| 66 |
+
jupyterlab-widgets==1.0.2
|
| 67 |
+
kiwisolver==1.3.2
|
| 68 |
+
lime==0.2.0.1
|
| 69 |
+
llvmlite==0.38.0
|
| 70 |
+
markdown==3.3.6
|
| 71 |
+
markupsafe==2.0.1
|
| 72 |
+
matplotlib==3.5.1
|
| 73 |
+
matplotlib-inline==0.1.3
|
| 74 |
+
mistune==0.8.4
|
| 75 |
+
multidict==6.0.2
|
| 76 |
+
multiprocess==0.70.12.2
|
| 77 |
+
mypy-extensions==0.4.3
|
| 78 |
+
nbclient==0.5.10
|
| 79 |
+
nbconvert==6.4.1
|
| 80 |
+
nbformat==5.1.3
|
| 81 |
+
nest-asyncio==1.5.4
|
| 82 |
+
networkx==2.6.3
|
| 83 |
+
nltk==3.6.7
|
| 84 |
+
notebook==6.4.8
|
| 85 |
+
numba==0.55.1
|
| 86 |
+
numpy==1.21.5
|
| 87 |
+
oauthlib==3.2.0
|
| 88 |
+
openpyxl==3.0.9
|
| 89 |
+
packaging==21.3
|
| 90 |
+
pandas==1.4.0
|
| 91 |
+
pandocfilters==1.5.0
|
| 92 |
+
parso==0.8.3
|
| 93 |
+
pathspec==0.9.0
|
| 94 |
+
pathtools==0.1.2
|
| 95 |
+
pexpect==4.8.0
|
| 96 |
+
pickleshare==0.7.5
|
| 97 |
+
pillow==9.0.0
|
| 98 |
+
platformdirs==2.4.1
|
| 99 |
+
plotly==5.5.0
|
| 100 |
+
prometheus-client==0.13.1
|
| 101 |
+
promise==2.3
|
| 102 |
+
prompt-toolkit==3.0.26
|
| 103 |
+
protobuf==3.19.4
|
| 104 |
+
psutil==5.9.0
|
| 105 |
+
ptyprocess==0.7.0
|
| 106 |
+
pure-eval==0.2.2
|
| 107 |
+
pyarrow==6.0.1
|
| 108 |
+
pyasn1==0.4.8
|
| 109 |
+
pyasn1-modules==0.2.8
|
| 110 |
+
pycparser==2.21
|
| 111 |
+
pydeck==0.7.1
|
| 112 |
+
pygments==2.11.2
|
| 113 |
+
pympler==1.0.1
|
| 114 |
+
pynndescent==0.5.6
|
| 115 |
+
pyparsing==3.0.7
|
| 116 |
+
pyrsistent==0.18.1
|
| 117 |
+
python-dateutil==2.8.2
|
| 118 |
+
pytz==2021.3
|
| 119 |
+
pytz-deprecation-shim==0.1.0.post0
|
| 120 |
+
pywavelets==1.2.0
|
| 121 |
+
pyyaml==6.0
|
| 122 |
+
pyzmq==22.3.0
|
| 123 |
+
regex==2022.1.18
|
| 124 |
+
requests==2.27.1
|
| 125 |
+
requests-oauthlib==1.3.1
|
| 126 |
+
rsa==4.8
|
| 127 |
+
sacremoses==0.0.47
|
| 128 |
+
scikit-image==0.19.1
|
| 129 |
+
scikit-learn==1.0.2
|
| 130 |
+
scikit-multilearn==0.2.0
|
| 131 |
+
scipy==1.7.3
|
| 132 |
+
semantic-version==2.8.5
|
| 133 |
+
send2trash==1.8.0
|
| 134 |
+
sentencepiece==0.1.96
|
| 135 |
+
sentry-sdk==1.5.4
|
| 136 |
+
seqeval==1.2.2
|
| 137 |
+
setuptools-rust==1.1.2
|
| 138 |
+
shortuuid==1.0.8
|
| 139 |
+
simpletransformers==0.63.4
|
| 140 |
+
six==1.16.0
|
| 141 |
+
smart-open==5.2.1
|
| 142 |
+
smmap==5.0.0
|
| 143 |
+
stack-data==0.1.4
|
| 144 |
+
streamlit==1.5.0
|
| 145 |
+
subprocess32==3.5.4
|
| 146 |
+
tenacity==8.0.1
|
| 147 |
+
tensorboard==2.8.0
|
| 148 |
+
tensorboard-data-server==0.6.1
|
| 149 |
+
tensorboard-plugin-wit==1.8.1
|
| 150 |
+
termcolor==1.1.0
|
| 151 |
+
terminado==0.13.1
|
| 152 |
+
testpath==0.5.0
|
| 153 |
+
threadpoolctl==3.1.0
|
| 154 |
+
tifffile==2021.11.2
|
| 155 |
+
tokenizers==0.11.4
|
| 156 |
+
toml==0.10.2
|
| 157 |
+
tomli==2.0.0
|
| 158 |
+
toolz==0.11.2
|
| 159 |
+
torch==1.9.0
|
| 160 |
+
torchvision==0.10.0
|
| 161 |
+
tornado==6.1
|
| 162 |
+
tqdm==4.62.3
|
| 163 |
+
traitlets==5.1.1
|
| 164 |
+
transformers==4.16.2
|
| 165 |
+
tzdata==2021.5
|
| 166 |
+
tzlocal==4.1
|
| 167 |
+
umap-learn==0.5.2
|
| 168 |
+
urllib3==1.26.8
|
| 169 |
+
validators==0.18.2
|
| 170 |
+
wandb==0.12.9
|
| 171 |
+
watchdog==2.1.6
|
| 172 |
+
wcwidth==0.2.5
|
| 173 |
+
webencodings==0.5.1
|
| 174 |
+
werkzeug==2.0.2
|
| 175 |
+
widgetsnbextension==3.5.2
|
| 176 |
+
xgboost==1.4.2
|
| 177 |
+
xxhash==2.0.2
|
| 178 |
+
yarl==1.7.2
|
| 179 |
+
yaspin==2.1.0
|
| 180 |
+
zipp==3.7.0
|