Spaces:
Sleeping
Sleeping
Upload core files
Browse files- .gitattributes +2 -35
- .gitignore +6 -0
- LICENSE +21 -0
- README.md +59 -13
- app.py +83 -0
- data/IRIS.csv +151 -0
- notebook.ipynb +0 -0
- requirements.txt +7 -0
- screenshots/App_Interface.png +0 -0
- src/Iris_App_toolkit +0 -0
- src/original_data_profile.html +0 -0
- src/xgb_model.json +0 -0
.gitattributes
CHANGED
|
@@ -1,35 +1,2 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
*.bin filter=lfs diff=lfs merge=lfs -text
|
| 4 |
-
*.bz2 filter=lfs diff=lfs merge=lfs -text
|
| 5 |
-
*.ckpt filter=lfs diff=lfs merge=lfs -text
|
| 6 |
-
*.ftz filter=lfs diff=lfs merge=lfs -text
|
| 7 |
-
*.gz filter=lfs diff=lfs merge=lfs -text
|
| 8 |
-
*.h5 filter=lfs diff=lfs merge=lfs -text
|
| 9 |
-
*.joblib filter=lfs diff=lfs merge=lfs -text
|
| 10 |
-
*.lfs.* filter=lfs diff=lfs merge=lfs -text
|
| 11 |
-
*.mlmodel filter=lfs diff=lfs merge=lfs -text
|
| 12 |
-
*.model filter=lfs diff=lfs merge=lfs -text
|
| 13 |
-
*.msgpack filter=lfs diff=lfs merge=lfs -text
|
| 14 |
-
*.npy filter=lfs diff=lfs merge=lfs -text
|
| 15 |
-
*.npz filter=lfs diff=lfs merge=lfs -text
|
| 16 |
-
*.onnx filter=lfs diff=lfs merge=lfs -text
|
| 17 |
-
*.ot filter=lfs diff=lfs merge=lfs -text
|
| 18 |
-
*.parquet filter=lfs diff=lfs merge=lfs -text
|
| 19 |
-
*.pb filter=lfs diff=lfs merge=lfs -text
|
| 20 |
-
*.pickle filter=lfs diff=lfs merge=lfs -text
|
| 21 |
-
*.pkl filter=lfs diff=lfs merge=lfs -text
|
| 22 |
-
*.pt filter=lfs diff=lfs merge=lfs -text
|
| 23 |
-
*.pth filter=lfs diff=lfs merge=lfs -text
|
| 24 |
-
*.rar filter=lfs diff=lfs merge=lfs -text
|
| 25 |
-
*.safetensors filter=lfs diff=lfs merge=lfs -text
|
| 26 |
-
saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
| 27 |
-
*.tar.* filter=lfs diff=lfs merge=lfs -text
|
| 28 |
-
*.tar filter=lfs diff=lfs merge=lfs -text
|
| 29 |
-
*.tflite filter=lfs diff=lfs merge=lfs -text
|
| 30 |
-
*.tgz filter=lfs diff=lfs merge=lfs -text
|
| 31 |
-
*.wasm filter=lfs diff=lfs merge=lfs -text
|
| 32 |
-
*.xz filter=lfs diff=lfs merge=lfs -text
|
| 33 |
-
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
-
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
-
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
| 1 |
+
# Auto detect text files and perform LF normalization
|
| 2 |
+
* text=auto
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
.gitignore
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
__pycache__
|
| 2 |
+
.ipynb_checkpoints
|
| 3 |
+
.vs
|
| 4 |
+
.vscode
|
| 5 |
+
catboost_info
|
| 6 |
+
venv
|
LICENSE
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
MIT License
|
| 2 |
+
|
| 3 |
+
Copyright (c) 2023 Kwame
|
| 4 |
+
|
| 5 |
+
Permission is hereby granted, free of charge, to any person obtaining a copy
|
| 6 |
+
of this software and associated documentation files (the "Software"), to deal
|
| 7 |
+
in the Software without restriction, including without limitation the rights
|
| 8 |
+
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
| 9 |
+
copies of the Software, and to permit persons to whom the Software is
|
| 10 |
+
furnished to do so, subject to the following conditions:
|
| 11 |
+
|
| 12 |
+
The above copyright notice and this permission notice shall be included in all
|
| 13 |
+
copies or substantial portions of the Software.
|
| 14 |
+
|
| 15 |
+
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
| 16 |
+
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
| 17 |
+
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
| 18 |
+
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
| 19 |
+
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
| 20 |
+
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
| 21 |
+
SOFTWARE.
|
README.md
CHANGED
|
@@ -1,13 +1,59 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
|
| 4 |
-
|
| 5 |
-
|
| 6 |
-
|
| 7 |
-
|
| 8 |
-
|
| 9 |
-
|
| 10 |
-
|
| 11 |
-
|
| 12 |
-
|
| 13 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Iris_Flower_Specie_Prediction
|
| 2 |
+
|
| 3 |
+
## Introduction
|
| 4 |
+
|
| 5 |
+
This project seeks to use machine learning to predict the species of an iris flower, given information on its petals and sepals. It also features an interface that makes it easier for users to interact with the model to predict the specie of an iris flower.
|
| 6 |
+
|
| 7 |
+
It comes as a challenge from the **Slightly Techie** community.
|
| 8 |
+
|
| 9 |
+
## Process Description
|
| 10 |
+
|
| 11 |
+
The process begins with exporting the necessary items from the notebook, building an interface that works correctly, importing the necessary items for modelling, and then writing the code to process inputs. The process can be summarized as:
|
| 12 |
+
|
| 13 |
+
- Export machine learning items from notebook,
|
| 14 |
+
- Import machine learning items into the app script,
|
| 15 |
+
- Build an interface,
|
| 16 |
+
- Write a function to process inputs,
|
| 17 |
+
- Pass values through the interface,
|
| 18 |
+
- Recover these values in backend,
|
| 19 |
+
- Apply the necessary processing,
|
| 20 |
+
- Submit the processed values to the ML model to make the predictions,
|
| 21 |
+
- Process the predictions obtained and display them on the interface.
|
| 22 |
+
|
| 23 |
+
## Installation
|
| 24 |
+
|
| 25 |
+
To setup and run this project you need to have [`Python3`](https://www.python.org/) installed on your system. Then you can clone this repo. At the repo's root, use the code from below which applies:
|
| 26 |
+
|
| 27 |
+
- Windows:
|
| 28 |
+
|
| 29 |
+
python -m venv venv; venv\Scripts\activate; python -m pip install -q --upgrade pip; python -m pip install -qr requirements.txt
|
| 30 |
+
|
| 31 |
+
- Linux & MacOs:
|
| 32 |
+
|
| 33 |
+
python3 -m venv venv; source venv/bin/activate; python -m pip install -q --upgrade pip; python -m pip install -qr requirements.txt
|
| 34 |
+
|
| 35 |
+
**NB:** For MacOs users, please install `Xcode` if you have an issue.
|
| 36 |
+
|
| 37 |
+
You can then run the app (still at the repository root):
|
| 38 |
+
|
| 39 |
+
- App built with Gradio Interface
|
| 40 |
+
|
| 41 |
+
python app.py
|
| 42 |
+
|
| 43 |
+
- With `inbrowser = True` defined, it should open a browser tab automatically. If it doesn't, type this address in your browser: <http://127.0.0.1:7860/>
|
| 44 |
+
|
| 45 |
+
## Screenshots
|
| 46 |
+
|
| 47 |
+
<table>
|
| 48 |
+
<tr>
|
| 49 |
+
<th>Gradio App Interface with a Prediction</th>
|
| 50 |
+
</tr>
|
| 51 |
+
<tr>
|
| 52 |
+
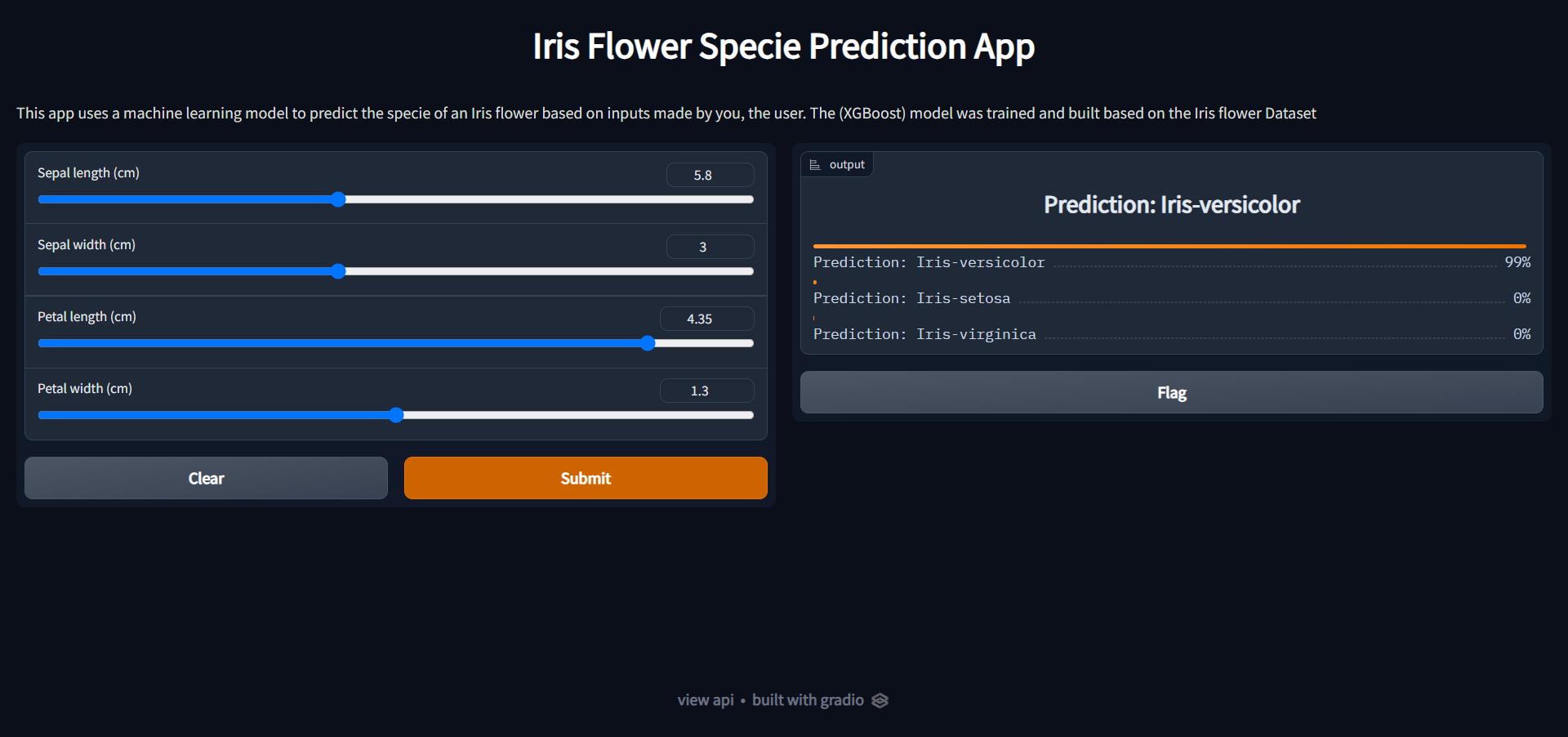
<td><img src="screenshots\App_Interface.png"/></td>
|
| 53 |
+
</tr>
|
| 54 |
+
</table>
|
| 55 |
+
|
| 56 |
+
## Contact Information
|
| 57 |
+
|
| 58 |
+
- [Kwame Otchere](https://kodoi-oj.github.io/)
|
| 59 |
+
- [](https://twitter.com/kwameoo_)
|
app.py
ADDED
|
@@ -0,0 +1,83 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# ----- Load base libraries and packages
|
| 2 |
+
import gradio as gr
|
| 3 |
+
|
| 4 |
+
import numpy as np
|
| 5 |
+
import pandas as pd
|
| 6 |
+
|
| 7 |
+
import os
|
| 8 |
+
import pickle
|
| 9 |
+
|
| 10 |
+
import xgboost as xgb
|
| 11 |
+
from xgboost import XGBClassifier
|
| 12 |
+
|
| 13 |
+
|
| 14 |
+
# ----- Useful lists
|
| 15 |
+
expected_inputs = ["sepal_length", "sepal_width", "petal_length", "petal_width"]
|
| 16 |
+
|
| 17 |
+
|
| 18 |
+
# ----- Helper Functions
|
| 19 |
+
# Function to load ML toolkit
|
| 20 |
+
def load_ml_toolkit(file_path= r"src\Iris_App_toolkit"):
|
| 21 |
+
"""
|
| 22 |
+
This function loads the ML items into this file. It takes the path to the ML items to load it.
|
| 23 |
+
|
| 24 |
+
Args:
|
| 25 |
+
file_path (regexp, optional): It receives the file path to the ML items, but defaults to the "src" folder in the repository. The full default relative path is r"src\Iris_App_toolkit".
|
| 26 |
+
|
| 27 |
+
Returns:
|
| 28 |
+
file: It returns the pickle file (which in this case contains the Machine Learning items.)
|
| 29 |
+
"""
|
| 30 |
+
|
| 31 |
+
with open(file_path, "rb") as file:
|
| 32 |
+
loaded_toolkit = pickle.load(file)
|
| 33 |
+
return loaded_toolkit
|
| 34 |
+
|
| 35 |
+
|
| 36 |
+
# Importing the toolkit
|
| 37 |
+
loaded_toolkit = load_ml_toolkit()
|
| 38 |
+
scaler = loaded_toolkit["scaler"]
|
| 39 |
+
|
| 40 |
+
# Import the model
|
| 41 |
+
model = XGBClassifier()
|
| 42 |
+
model.load_model(r"src\xgb_model.json")
|
| 43 |
+
|
| 44 |
+
|
| 45 |
+
# Function to process inputs and return prediction
|
| 46 |
+
def process_and_predict(*args, scaler=scaler, model=model):
|
| 47 |
+
"""
|
| 48 |
+
This function processes the inputs and returns the predicted specie of the flower
|
| 49 |
+
It receives the user inputs, scaler and model. The inputs are then put through the same process as was done during modelling
|
| 50 |
+
|
| 51 |
+
Args:
|
| 52 |
+
scaler (MinMaxScaler, optional): It is the scaler (MinMaxScaler) used to scale the numeric features before training the model, and should be loaded either as part of the ML Items or as a standalone item. Defaults to scaler, which comes with the ML Items dictionary.
|
| 53 |
+
model (XGBoost, optional): This is the model that was trained and is to be used for the prediction. Since XGBoost seems to have issues with Pickle, import as a standalone. It defaults to "model", as loaded.
|
| 54 |
+
|
| 55 |
+
Returns:
|
| 56 |
+
Prediction (label): Returns the label of the predicted class, i.e. the specie of the flower
|
| 57 |
+
"""
|
| 58 |
+
|
| 59 |
+
# Convert inputs into a DataFrame
|
| 60 |
+
input_data = pd.DataFrame([args], columns=expected_inputs)
|
| 61 |
+
|
| 62 |
+
# Scale the numeric columns
|
| 63 |
+
input_data[expected_inputs] = scaler.transform(input_data[expected_inputs])
|
| 64 |
+
|
| 65 |
+
# Make the prediction
|
| 66 |
+
model_output = model.predict_proba(input_data)
|
| 67 |
+
setosa_prob = float(model_output[0][0])
|
| 68 |
+
versicolor_prob = float(model_output[0][1])
|
| 69 |
+
virginica_prob = 1 - (setosa_prob + versicolor_prob)
|
| 70 |
+
return {"Prediction: Iris-setosa": setosa_prob, "Prediction: Iris-versicolor": versicolor_prob, "Prediction: Iris-virginica": virginica_prob}
|
| 71 |
+
|
| 72 |
+
|
| 73 |
+
# ----- App Interface
|
| 74 |
+
# Inputs
|
| 75 |
+
sepal_length = gr.Slider(label="Sepal length (cm)", minimum=4.3, step=0.1, maximum= 7.9, interactive=True, value=5.8)
|
| 76 |
+
sepal_width = gr.Slider(label="Sepal width (cm)", minimum=2, step=0.1, maximum= 4.4, interactive=True, value=3)
|
| 77 |
+
petal_length = gr.Slider(label="Petal length (cm)", minimum=1, step=0.05, maximum= 4.9, interactive=True, value=4.35)
|
| 78 |
+
petal_width = gr.Slider(label="Petal width (cm)", minimum=0.1, step=0.05, maximum= 2.5, interactive=True, value=1.3)
|
| 79 |
+
|
| 80 |
+
|
| 81 |
+
# Output
|
| 82 |
+
gr.Interface(inputs=[sepal_length, sepal_width, petal_length, petal_width], outputs=gr.Label("Awaiting Submission..."), fn=process_and_predict, title="Iris Flower Specie Prediction App",
|
| 83 |
+
description="""This app uses a machine learning model to predict the specie of an Iris flower based on inputs made by you, the user. The (XGBoost) model was trained and built based on the Iris flower Dataset""").launch(inbrowser=True, show_error=True)
|
data/IRIS.csv
ADDED
|
@@ -0,0 +1,151 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
sepal_length,sepal_width,petal_length,petal_width,species
|
| 2 |
+
5.1,3.5,1.4,0.2,Iris-setosa
|
| 3 |
+
4.9,3,1.4,0.2,Iris-setosa
|
| 4 |
+
4.7,3.2,1.3,0.2,Iris-setosa
|
| 5 |
+
4.6,3.1,1.5,0.2,Iris-setosa
|
| 6 |
+
5,3.6,1.4,0.2,Iris-setosa
|
| 7 |
+
5.4,3.9,1.7,0.4,Iris-setosa
|
| 8 |
+
4.6,3.4,1.4,0.3,Iris-setosa
|
| 9 |
+
5,3.4,1.5,0.2,Iris-setosa
|
| 10 |
+
4.4,2.9,1.4,0.2,Iris-setosa
|
| 11 |
+
4.9,3.1,1.5,0.1,Iris-setosa
|
| 12 |
+
5.4,3.7,1.5,0.2,Iris-setosa
|
| 13 |
+
4.8,3.4,1.6,0.2,Iris-setosa
|
| 14 |
+
4.8,3,1.4,0.1,Iris-setosa
|
| 15 |
+
4.3,3,1.1,0.1,Iris-setosa
|
| 16 |
+
5.8,4,1.2,0.2,Iris-setosa
|
| 17 |
+
5.7,4.4,1.5,0.4,Iris-setosa
|
| 18 |
+
5.4,3.9,1.3,0.4,Iris-setosa
|
| 19 |
+
5.1,3.5,1.4,0.3,Iris-setosa
|
| 20 |
+
5.7,3.8,1.7,0.3,Iris-setosa
|
| 21 |
+
5.1,3.8,1.5,0.3,Iris-setosa
|
| 22 |
+
5.4,3.4,1.7,0.2,Iris-setosa
|
| 23 |
+
5.1,3.7,1.5,0.4,Iris-setosa
|
| 24 |
+
4.6,3.6,1,0.2,Iris-setosa
|
| 25 |
+
5.1,3.3,1.7,0.5,Iris-setosa
|
| 26 |
+
4.8,3.4,1.9,0.2,Iris-setosa
|
| 27 |
+
5,3,1.6,0.2,Iris-setosa
|
| 28 |
+
5,3.4,1.6,0.4,Iris-setosa
|
| 29 |
+
5.2,3.5,1.5,0.2,Iris-setosa
|
| 30 |
+
5.2,3.4,1.4,0.2,Iris-setosa
|
| 31 |
+
4.7,3.2,1.6,0.2,Iris-setosa
|
| 32 |
+
4.8,3.1,1.6,0.2,Iris-setosa
|
| 33 |
+
5.4,3.4,1.5,0.4,Iris-setosa
|
| 34 |
+
5.2,4.1,1.5,0.1,Iris-setosa
|
| 35 |
+
5.5,4.2,1.4,0.2,Iris-setosa
|
| 36 |
+
4.9,3.1,1.5,0.1,Iris-setosa
|
| 37 |
+
5,3.2,1.2,0.2,Iris-setosa
|
| 38 |
+
5.5,3.5,1.3,0.2,Iris-setosa
|
| 39 |
+
4.9,3.1,1.5,0.1,Iris-setosa
|
| 40 |
+
4.4,3,1.3,0.2,Iris-setosa
|
| 41 |
+
5.1,3.4,1.5,0.2,Iris-setosa
|
| 42 |
+
5,3.5,1.3,0.3,Iris-setosa
|
| 43 |
+
4.5,2.3,1.3,0.3,Iris-setosa
|
| 44 |
+
4.4,3.2,1.3,0.2,Iris-setosa
|
| 45 |
+
5,3.5,1.6,0.6,Iris-setosa
|
| 46 |
+
5.1,3.8,1.9,0.4,Iris-setosa
|
| 47 |
+
4.8,3,1.4,0.3,Iris-setosa
|
| 48 |
+
5.1,3.8,1.6,0.2,Iris-setosa
|
| 49 |
+
4.6,3.2,1.4,0.2,Iris-setosa
|
| 50 |
+
5.3,3.7,1.5,0.2,Iris-setosa
|
| 51 |
+
5,3.3,1.4,0.2,Iris-setosa
|
| 52 |
+
7,3.2,4.7,1.4,Iris-versicolor
|
| 53 |
+
6.4,3.2,4.5,1.5,Iris-versicolor
|
| 54 |
+
6.9,3.1,4.9,1.5,Iris-versicolor
|
| 55 |
+
5.5,2.3,4,1.3,Iris-versicolor
|
| 56 |
+
6.5,2.8,4.6,1.5,Iris-versicolor
|
| 57 |
+
5.7,2.8,4.5,1.3,Iris-versicolor
|
| 58 |
+
6.3,3.3,4.7,1.6,Iris-versicolor
|
| 59 |
+
4.9,2.4,3.3,1,Iris-versicolor
|
| 60 |
+
6.6,2.9,4.6,1.3,Iris-versicolor
|
| 61 |
+
5.2,2.7,3.9,1.4,Iris-versicolor
|
| 62 |
+
5,2,3.5,1,Iris-versicolor
|
| 63 |
+
5.9,3,4.2,1.5,Iris-versicolor
|
| 64 |
+
6,2.2,4,1,Iris-versicolor
|
| 65 |
+
6.1,2.9,4.7,1.4,Iris-versicolor
|
| 66 |
+
5.6,2.9,3.6,1.3,Iris-versicolor
|
| 67 |
+
6.7,3.1,4.4,1.4,Iris-versicolor
|
| 68 |
+
5.6,3,4.5,1.5,Iris-versicolor
|
| 69 |
+
5.8,2.7,4.1,1,Iris-versicolor
|
| 70 |
+
6.2,2.2,4.5,1.5,Iris-versicolor
|
| 71 |
+
5.6,2.5,3.9,1.1,Iris-versicolor
|
| 72 |
+
5.9,3.2,4.8,1.8,Iris-versicolor
|
| 73 |
+
6.1,2.8,4,1.3,Iris-versicolor
|
| 74 |
+
6.3,2.5,4.9,1.5,Iris-versicolor
|
| 75 |
+
6.1,2.8,4.7,1.2,Iris-versicolor
|
| 76 |
+
6.4,2.9,4.3,1.3,Iris-versicolor
|
| 77 |
+
6.6,3,4.4,1.4,Iris-versicolor
|
| 78 |
+
6.8,2.8,4.8,1.4,Iris-versicolor
|
| 79 |
+
6.7,3,5,1.7,Iris-versicolor
|
| 80 |
+
6,2.9,4.5,1.5,Iris-versicolor
|
| 81 |
+
5.7,2.6,3.5,1,Iris-versicolor
|
| 82 |
+
5.5,2.4,3.8,1.1,Iris-versicolor
|
| 83 |
+
5.5,2.4,3.7,1,Iris-versicolor
|
| 84 |
+
5.8,2.7,3.9,1.2,Iris-versicolor
|
| 85 |
+
6,2.7,5.1,1.6,Iris-versicolor
|
| 86 |
+
5.4,3,4.5,1.5,Iris-versicolor
|
| 87 |
+
6,3.4,4.5,1.6,Iris-versicolor
|
| 88 |
+
6.7,3.1,4.7,1.5,Iris-versicolor
|
| 89 |
+
6.3,2.3,4.4,1.3,Iris-versicolor
|
| 90 |
+
5.6,3,4.1,1.3,Iris-versicolor
|
| 91 |
+
5.5,2.5,4,1.3,Iris-versicolor
|
| 92 |
+
5.5,2.6,4.4,1.2,Iris-versicolor
|
| 93 |
+
6.1,3,4.6,1.4,Iris-versicolor
|
| 94 |
+
5.8,2.6,4,1.2,Iris-versicolor
|
| 95 |
+
5,2.3,3.3,1,Iris-versicolor
|
| 96 |
+
5.6,2.7,4.2,1.3,Iris-versicolor
|
| 97 |
+
5.7,3,4.2,1.2,Iris-versicolor
|
| 98 |
+
5.7,2.9,4.2,1.3,Iris-versicolor
|
| 99 |
+
6.2,2.9,4.3,1.3,Iris-versicolor
|
| 100 |
+
5.1,2.5,3,1.1,Iris-versicolor
|
| 101 |
+
5.7,2.8,4.1,1.3,Iris-versicolor
|
| 102 |
+
6.3,3.3,6,2.5,Iris-virginica
|
| 103 |
+
5.8,2.7,5.1,1.9,Iris-virginica
|
| 104 |
+
7.1,3,5.9,2.1,Iris-virginica
|
| 105 |
+
6.3,2.9,5.6,1.8,Iris-virginica
|
| 106 |
+
6.5,3,5.8,2.2,Iris-virginica
|
| 107 |
+
7.6,3,6.6,2.1,Iris-virginica
|
| 108 |
+
4.9,2.5,4.5,1.7,Iris-virginica
|
| 109 |
+
7.3,2.9,6.3,1.8,Iris-virginica
|
| 110 |
+
6.7,2.5,5.8,1.8,Iris-virginica
|
| 111 |
+
7.2,3.6,6.1,2.5,Iris-virginica
|
| 112 |
+
6.5,3.2,5.1,2,Iris-virginica
|
| 113 |
+
6.4,2.7,5.3,1.9,Iris-virginica
|
| 114 |
+
6.8,3,5.5,2.1,Iris-virginica
|
| 115 |
+
5.7,2.5,5,2,Iris-virginica
|
| 116 |
+
5.8,2.8,5.1,2.4,Iris-virginica
|
| 117 |
+
6.4,3.2,5.3,2.3,Iris-virginica
|
| 118 |
+
6.5,3,5.5,1.8,Iris-virginica
|
| 119 |
+
7.7,3.8,6.7,2.2,Iris-virginica
|
| 120 |
+
7.7,2.6,6.9,2.3,Iris-virginica
|
| 121 |
+
6,2.2,5,1.5,Iris-virginica
|
| 122 |
+
6.9,3.2,5.7,2.3,Iris-virginica
|
| 123 |
+
5.6,2.8,4.9,2,Iris-virginica
|
| 124 |
+
7.7,2.8,6.7,2,Iris-virginica
|
| 125 |
+
6.3,2.7,4.9,1.8,Iris-virginica
|
| 126 |
+
6.7,3.3,5.7,2.1,Iris-virginica
|
| 127 |
+
7.2,3.2,6,1.8,Iris-virginica
|
| 128 |
+
6.2,2.8,4.8,1.8,Iris-virginica
|
| 129 |
+
6.1,3,4.9,1.8,Iris-virginica
|
| 130 |
+
6.4,2.8,5.6,2.1,Iris-virginica
|
| 131 |
+
7.2,3,5.8,1.6,Iris-virginica
|
| 132 |
+
7.4,2.8,6.1,1.9,Iris-virginica
|
| 133 |
+
7.9,3.8,6.4,2,Iris-virginica
|
| 134 |
+
6.4,2.8,5.6,2.2,Iris-virginica
|
| 135 |
+
6.3,2.8,5.1,1.5,Iris-virginica
|
| 136 |
+
6.1,2.6,5.6,1.4,Iris-virginica
|
| 137 |
+
7.7,3,6.1,2.3,Iris-virginica
|
| 138 |
+
6.3,3.4,5.6,2.4,Iris-virginica
|
| 139 |
+
6.4,3.1,5.5,1.8,Iris-virginica
|
| 140 |
+
6,3,4.8,1.8,Iris-virginica
|
| 141 |
+
6.9,3.1,5.4,2.1,Iris-virginica
|
| 142 |
+
6.7,3.1,5.6,2.4,Iris-virginica
|
| 143 |
+
6.9,3.1,5.1,2.3,Iris-virginica
|
| 144 |
+
5.8,2.7,5.1,1.9,Iris-virginica
|
| 145 |
+
6.8,3.2,5.9,2.3,Iris-virginica
|
| 146 |
+
6.7,3.3,5.7,2.5,Iris-virginica
|
| 147 |
+
6.7,3,5.2,2.3,Iris-virginica
|
| 148 |
+
6.3,2.5,5,1.9,Iris-virginica
|
| 149 |
+
6.5,3,5.2,2,Iris-virginica
|
| 150 |
+
6.2,3.4,5.4,2.3,Iris-virginica
|
| 151 |
+
5.9,3,5.1,1.8,Iris-virginica
|
notebook.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
requirements.txt
ADDED
|
@@ -0,0 +1,7 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
gradio
|
| 2 |
+
numpy==1.23.5
|
| 3 |
+
pandas==1.5.2
|
| 4 |
+
seaborn==0.12.2
|
| 5 |
+
xgboost==1.7.3
|
| 6 |
+
lightgbm==3.2.1
|
| 7 |
+
sweetviz==2.1.4
|
screenshots/App_Interface.png
ADDED
|
src/Iris_App_toolkit
ADDED
|
Binary file (237 kB). View file
|
|
|
src/original_data_profile.html
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
src/xgb_model.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|